Resolving Identifiability Issues in Mathematical Models of Inflammation: A Guide for Robust Quantification
Mathematical modeling is crucial for understanding the complex dynamics of inflammatory responses and developing effective therapeutics.
Resolving Identifiability Issues in Mathematical Models of Inflammation: A Guide for Robust Quantification
Abstract
Mathematical modeling is crucial for understanding the complex dynamics of inflammatory responses and developing effective therapeutics. However, the utility of these models is often compromised by identifiability issues, where model parameters cannot be uniquely or reliably estimated from available data. This article provides a comprehensive guide for researchers and drug development professionals on addressing these critical challenges. We explore the foundational concepts of structural and practical identifiability, review advanced methodological and computational tools for analysis, present strategies for troubleshooting and optimizing non-identifiable models, and discuss rigorous validation frameworks. By synthesizing the latest research, this article serves as a practical resource for developing more reliable, predictive models of inflammation to enhance drug discovery and personalized medicine approaches.
Understanding Identifiability: Why Your Inflammation Model's Parameters Might Be Unknowable
Frequently Asked Questions (FAQs)
FAQ 1: What is the fundamental difference between structural and practical identifiability?
- Answer: Structural identifiability is a theoretical property of your model's equations. It assesses whether parameters can be uniquely determined from perfect, continuous, and noise-free data. It is a necessary prerequisite for reliable parameter estimation [1] [2] [3]. Practical identifiability, in contrast, concerns whether parameters can be uniquely estimated given your actual data—which is finite, noisy, and collected at specific time points [4] [5]. Even a structurally identifiable model can be practically unidentifiable if the data is insufficient or too noisy to reliably estimate the parameters [6] [5].
FAQ 2: Why should I perform identifiability analysis before fitting my model to experimental data?
- Answer: Conducting identifiability analysis before experiments helps you avoid building models or designing experiments that are fundamentally incapable of providing unique parameter estimates. It saves significant time and resources by [2]:
- Revealing if your model structure is over-parameterized.
- Guiding the selection of measurable outputs.
- Informing experimental design (e.g., when to take samples) to ensure the data collected will be informative.
- Answer: Conducting identifiability analysis before experiments helps you avoid building models or designing experiments that are fundamentally incapable of providing unique parameter estimates. It saves significant time and resources by [2]:
FAQ 3: A parameter in my model is structurally unidentifiable. What are my options?
- Answer: You have several troubleshooting paths:
- Reparameterize the model: Combine unidentifiable parameters into an identifiable composite parameter [2] [6].
- Fix parameter values: If possible, use literature values to set unidentifiable parameters to a constant value [2].
- Modify the model output: Sometimes, measuring an additional variable (e.g., another cytokine in an inflammation model) can render previously unidentifiable parameters identifiable [2].
- Simplify the model: Remove the mechanistic parts that cause unidentifiability if they are not critical to your research question.
- Answer: You have several troubleshooting paths:
FAQ 4: My model is structurally identifiable, but parameters are practically unidentifiable. How can I improve this?
- Answer: Practical unidentifiability is often addressed by improving the data or the estimation process [6] [5]:
- Optimal Experimental Design: Design experiments to maximize the information content in your data, for instance, by sampling during dynamic transition phases rather than at steady state [5].
- Increase Data Quality and Quantity: Collect more data points or reduce measurement noise.
- Use Regularization: Incorporate penalties (e.g., L2 regularization) during estimation to constrain parameter values [6].
- Answer: Practical unidentifiability is often addressed by improving the data or the estimation process [6] [5]:
Troubleshooting Guide: Diagnosing Identifiability Problems
Use this guide to diagnose and resolve common identifiability issues in mathematical modeling.
| Symptom | Likely Cause | Diagnostic Tools | Potential Solutions |
|---|---|---|---|
| Large confidence intervals for parameter estimates; small parameter changes drastically worsen fit [5]. | Practical Unidentifiability: Noisy or insufficient data, poor experimental design. | Profile Likelihood [1] [4], Monte Carlo simulations [7] [5], Fisher Information Matrix (FIM) analysis [4] [5]. | Optimal experimental design [5], collect more informative data, use regularization [6]. |
| Parameter estimates change drastically with different initial guesses; optimization fails to converge. | Structural or Practical Unidentifiability. | Structural identifiability tools (e.g., DAISY, StructuralIdentifiability.jl [4] [3] [7]), Profile Likelihood [1]. | First, confirm structural identifiability. If structurally identifiable, see solutions for practical unidentifiability. |
| Strong correlations between different parameter estimates. | Structural Unidentifiability or near-unidentifiability; parameters exist in a sloppy combination [2]. | Correlation matrix analysis, sensitivity analysis [8] [9], FIM eigenvalue decomposition (near-zero eigenvalues) [4] [5]. | Model reparameterization (combine parameters) [2], fix one of the correlated parameters from literature. |
| Good model fit (low error) but biologically implausible parameter values. | Structural Unidentifiability: Multiple parameter sets yield identical output [2]. | Structural identifiability analysis (e.g., Taylor series, EAR approach [2]). | Redesign model structure, impose biologically plausible constraints during estimation, measure additional model outputs. |
Experimental Protocols for Identifiability Analysis
Protocol 1: Profile Likelihood Analysis for Practical Identifiability
This protocol assesses how well a parameter can be identified from a given dataset by exploring the likelihood surface [1] [4].
- Define the Objective Function: Establish a cost function, such as the negative log-likelihood or sum of squared errors, between your model simulation and experimental data.
- Obtain Point Estimate: Find the parameter values that minimize the cost function. This is your best-fit estimate, ( \hat{\theta} ).
- Profile a Parameter: Select a parameter of interest, ( \thetai ). Over a defined range of values for ( \thetai ), for each fixed value, re-optimize the cost function over all other parameters ( \theta_{j \neq i} ).
- Plot and Interpret: Plot the optimized cost function value against the values of ( \theta_i ). A uniquely identifiable parameter will show a sharply peaked profile. A flat or shallow profile indicates practical unidentifiability [1] [5].
Protocol 2: Structural Identifiability Analysis using the Taylor Series Method
This algebraic method checks if the model's output is unique for all possible parameter values, assuming perfect data [2].
- Model Formulation: Express your model in the standard state-space form (ODE system with specified outputs) [2].
- Taylor Series Expansion: Expand the model output ( y(t) ) as a Taylor series around a known time point (typically ( t=0 )), using the model's equations to compute higher-order derivatives.
- Coefficient Comparison: The coefficients of the Taylor series ( (y(0), y'(0), y''(0), \dots) ) are functions of the unknown parameters and initial conditions. The model is structurally globally identifiable if these equations can be solved uniquely for all parameters. If the solution is unique only in a local neighborhood, it is locally identifiable [2].
Signaling Pathways and Workflows
Identifiability Analysis Workflow
This diagram outlines the logical sequence for diagnosing and resolving identifiability issues in model development.
Inflammation Model Component Interactions
This diagram illustrates the core interactions in a typical cytokine-mediated inflammation model, a key application area where identifiability is crucial [8] [9].
The Scientist's Toolkit: Research Reagent Solutions
Table: Key computational tools and methods for identifiability analysis.
| Tool / Method | Function | Application Context |
|---|---|---|
| Profile Likelihood [1] [3] | Assesses practical identifiability by exploring likelihood-based confidence intervals for parameters. | ODE/PDE models; requires a defined cost function and optimization routine. |
| DAISY [4] | Performs structural identifiability analysis using differential algebra. Provides a categorical (yes/no) answer. | Models described by systems of rational ODEs; assumes perfect data. |
| StructuralIdentifiability.jl [3] [7] | A Julia library for assessing structural identifiability using a differential algebra approach. | Handles nonlinear ODE models; useful for complex biological systems. |
| Fisher Information Matrix (FIM) [4] [5] | A matrix whose inverse lower-bounds the covariance of parameters. Near-zero eigenvalues indicate unidentifiable directions. | Local, practical identifiability; requires parameter sensitivities. |
| Sensitivity Matrix Method (SMM) [4] | Analyzes the matrix of output sensitivities to parameters. A non-trivial null space indicates unidentifiability. | Practical identifiability; helps identify correlated parameters. |
| Monte Carlo Simulations [7] [5] | Evaluates practical identifiability by simulating noisy data and assessing the distribution of parameter estimates. | Quantifies robustness of parameter estimation to observational noise. |
| Oxaprotiline | Oxaprotiline, CAS:56433-44-4, MF:C20H23NO, MW:293.4 g/mol | Chemical Reagent |
| Picrotoxinin | Picrotoxinin, CAS:17617-45-7, MF:C15H16O6, MW:292.28 g/mol | Chemical Reagent |
The Critical Role of Identifiability in Predictive Immunology
In the field of predictive immunology, mathematical models are indispensable for interpreting complex biological processes, from viral infection dynamics to inflammatory resolution. Identifiability is a fundamental property that determines whether the parameters of a mathematical model can be uniquely estimated from available experimental data [10] [11]. The failure to ensure identifiability can lead to misleading parameter estimates, unreliable biological interpretations, and ultimately, flawed public health or therapeutic recommendations [11] [12].
This technical support center addresses the critical identifiability challenges faced by researchers in mathematical immunology. The guidance herein is framed within the broader thesis that resolving identifiability issues is paramount for developing models with genuine predictive power, particularly in the context of inflammation research.
FAQs: Core Concepts of Identifiability
Q1: What is the difference between structural and practical identifiability?
- Structural Identifiability is a theoretical property of the model itself. A model is structurally identifiable if, given perfect and noise-free experimental data, its parameters can be uniquely determined. This depends on the model's structure, the choice of observables, and the initial conditions of the state variables [10].
- Practical Identifiability refers to the ability to uniquely estimate parameters from real-world data that is typically sparse, noisy, and limited. Even a structurally identifiable model may not be practically identifiable if the available data is insufficient to constrain the parameters [10] [12].
Q2: Why is identifiability analysis crucial for mathematical models of inflammation?
Identifiability analysis is critical because models of immune processes, such as acute infection development or inflammatory resolution, often contain numerous parameters [10] [12]. Without verifying identifiability, researchers risk:
- Drawing incorrect conclusions about underlying biological mechanisms.
- Making unreliable predictions about disease outcomes or therapeutic efficacy.
- Developing models that are over-parameterized and lack predictive value [11] [12].
Q3: What common factors cause identifiability issues in immunological models?
- Model Over-parameterization: Using models with too many parameters relative to the available data [12].
- Inadequate Data: Using data that is sparse in time, has high noise, or lacks critical observables [10] [11] [12].
- Parameter Correlation: Strong dependencies between parameters, such as between the community transmission rate, the fraction of under-reporting, and the proportion of the population with prior immunity, which can make them impossible to estimate jointly from case data alone [11].
- Poor Experimental Design: Initial conditions and measurement protocols that do not provide sufficient information to tease apart parameter values [10].
Q4: How can I resolve the unidentifiability of key parameters in an epidemic model?
A common identifiability problem involves jointly estimating the transmission rate, under-reporting fraction, and prior immunity level from only reported case data, which is often unidentifiable [11]. This can be resolved by complementing the case data with additional information sources. Research shows that identifiability of all three parameters is achieved if reported incidence is complemented with sample survey data of prior immunity or prevalence during the outbreak [11].
Troubleshooting Guides
Guide 1: Addressing Structural Unidentifiability
Table 1: Strategies to Overcome Structural Unidentifiability
| Problem | Diagnostic Signs | Solution | Protocol/Method |
|---|---|---|---|
| Parameter Correlation | Parameters cannot be uniquely estimated even with perfect data; profiles are flat [11]. | Reformulate the model or reduce the number of parameters. | Use the profile likelihood approach to detect flat profiles. Fix one correlated parameter to a literature value to test the identifiability of others [11] [12]. |
| Insufficient Observables | Key state variables of the model (e.g., specific immune cell counts) are not measured [10]. | Increase the number of measured outputs. | Design experiments to measure additional model variables. For example, in viral infection models, measure both viral load and Cytotoxic T Lymphocyte (CTL) response kinetics [10]. |
| Complex Model Terms | A model with simple bilinear terms (e.g., for virus-CTL interactions) is not identifiable [10]. | Reparameterize the model with biologically realistic, bounded terms. | Refine bilinear terms to bounded-rate parameterizations, such as Michaelis-Menten-type functions, which can improve structural identifiability [10]. |
Guide 2: Addressing Practical Unidentifiability
Table 2: Strategies to Overcome Practical Unidentifiability
| Problem | Diagnostic Signs | Solution | Protocol/Method |
|---|---|---|---|
| Noisy or Sparse Data | Wide confidence intervals for parameter estimates; estimates vary significantly with different data realizations [12]. | Improve data quality and quantity. | Increase the frequency and precision of sampling. Use Bayesian estimation approaches with informative priors where justified to constrain parameter space [10]. |
| Poor Initial Conditions | Parameter estimates are highly sensitive to the initial guess for state variables [10] [12]. | Better initial state determination. | Perform rigorous initial state estimation or design experiments to directly measure initial conditions where possible [10]. |
| Inadequate Data Types | Case data alone is insufficient to identify all parameters of interest [11]. | Integrate multiple data sources. | Combine time-series data (e.g., incidence) with cross-sectional data (e.g., serological surveys for prior immunity or prevalence data) [11]. |
Experimental Protocols for Identifiability Analysis
Protocol 1: Structural Identifiability Analysis using Differential Algebra
Purpose: To determine if a system of Ordinary Differential Equations (ODEs) is structurally identifiable. Reagents & Tools: Computer with Julia programming environment, StructuralIdentifiability.jl package [10]. Workflow:
- Model Formulation: Define your ODE model, specifying state variables, parameters, and output functions (observables).
- Software Implementation: Code the model into the StructuralIdentifiability.jl package.
- Analysis Execution: Run the package's algorithm (e.g., based on differential algebra or generating series) to obtain identifiability results.
- Result Interpretation: The output will classify each parameter as either "globally identifiable," "locally identifiable," or "unidentifiable."
Structural identifiability analysis workflow.
Protocol 2: Practical Identifiability and Parameter Estimation using Bayesian Methods
Purpose: To estimate model parameters from noisy data and assess their practical identifiability by examining posterior distributions. Reagents & Tools: Computer with Julia/Python/R, DynamicHMC.jl package (or similar MCMC toolbox) [10]. Workflow:
- Data Preparation: Collect and preprocess experimental data (e.g., viral load and CTL response kinetics over time) [10].
- Model Definition: Use a structurally identifiable model.
- Bayesian Estimation: Implement a Hamiltonian Monte Carlo (HMC) sampler, such as the No-U-Turn Sampler (NUTS), to sample from the posterior distribution of the parameters [10].
- Diagnostic Checks: Analyze the Markov Chain Monte Carlo (MCMC) chains for convergence. Examine the posterior distributions: well-defined, peaked distributions indicate practical identifiability, while flat or multi-modal distributions suggest unidentifiability.
Practical identifiability analysis with Bayesian methods.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Tools for Modeling and Analysis in Immunology
| Item/Tool | Function/Application | Example/Note |
|---|---|---|
| StructuralIdentifiability.jl | A Julia-based package for analyzing the structural identifiability of ODE models. | Used to prove identifiability before costly parameter estimation exercises [10]. |
| DynamicHMC.jl | A Julia package for Bayesian inference using Hamiltonian Monte Carlo (HMC). | Enables robust parameter estimation and practical identifiability assessment via posterior distributions [10]. |
| Multiplex Immunoassays | Simultaneously measure concentrations of multiple soluble immune factors (cytokines, chemokines) from serum. | Critical for collecting rich, multi-dimensional data for model fitting (e.g., ProcartaPlex Human Inflammation Panel) [13]. |
| Profile Likelihood | A numerical method for investigating practical identifiability and confidence intervals. | Can reveal parameter correlations and unidentifiability that might be missed by other methods [12]. |
| SIAN (Software for Structural Identifiability Analysis) | Another software tool for structural identifiability analysis of ODE models. | An alternative to StructuralIdentifiability.jl [10]. |
| Naftazone | Naftazone CAS 15687-37-3|Research Chemical | Naftazone, a naphthoquinone derivative for vascular disease research. For Research Use Only. Not for human or veterinary use. |
| Pipobroman | Pipobroman, CAS:54-91-1, MF:C10H16Br2N2O2, MW:356.05 g/mol | Chemical Reagent |
Troubleshooting Guide: Frequently Asked Questions
1. What is the fundamental difference between structural and practical non-identifiability?
Structural non-identifiability is an inherent property of your model structure, where multiple parameter sets produce identical model outputs even with perfect, continuous, noise-free data. This occurs due to parameter correlations built into the model equations themselves [4]. In contrast, practical non-identifiability arises from limitations in your experimental data—such as sparse sampling times, significant measurement noise, or insufficient data points—which prevent unique parameter estimation despite the model being structurally identifiable [14] [15].
2. How can I detect non-identifiability in my inflammation model?
You can employ several methodological approaches. Collinearity analysis examines parameter correlations by calculating a collinearity index; high values (typically >10-15) indicate strong correlations and potential non-identifiability [16]. Likelihood profiling analyzes the flatness of likelihood curves for each parameter; flat profiles suggest the parameter cannot be uniquely identified [16] [17]. Structural identifiability tools like DAISY or StructuralIdentifiability.jl use differential algebra to provide definitive answers about structural identifiability for ordinary differential equation models [18] [4].
3. Why does my inflammation model with many parameters often become non-identifiable?
Complex inflammation models frequently incorporate numerous poorly constrained parameters while being calibrated against limited experimental data (e.g., only cytokine concentrations). This creates a situation where insufficient calibration targets relative to unknown parameters allows multiple parameter combinations to fit the same data equally well. This is particularly problematic in within-host pathogen models and physiological models of systemic inflammation [16] [14] [19].
4. What are the practical consequences of non-identifiability for drug development?
Non-identifiability can significantly impact decision-making in pharmaceutical development. Different, equally well-fitting parameter sets may produce divergent predictions about treatment effectiveness. For example, one study demonstrated that two different parameter sets fitting the same calibration targets yielded substantially different estimates of treatment benefit (0.67 vs. 0.31 life-years gained), potentially leading to incorrect decisions about treatment prioritization [16].
5. Can I still use a non-identifiable model for predictions?
Yes, but with important caveats. While a non-identifiable model may reliably predict the specific variables it was calibrated against, its predictions for unmeasured variables or different experimental conditions may be highly unreliable [15]. The model's predictive power for a particular variable depends on whether that variable was included in the training data, with successively adding more measured variables improving overall predictive capability [15].
Detection Methods for Non-Identifiability
Table 1: Comparison of Identifiability Analysis Methods
| Method | Type of Identifiability Assessed | Key Principle | Software Tools | Best Use Cases |
|---|---|---|---|---|
| Differential Algebra | Structural | Symbolic computation to eliminate unobserved variables | DAISY, StructuralIdentifiability.jl [18] [4] | A priori analysis of model structure |
| Profile Likelihood | Practical | Examination of parameter likelihood profiles | Custom implementation in MATLAB/R/Python [16] [17] | Assessing identifiability with existing datasets |
| Fisher Information Matrix | Practical | Analysis of curvature in parameter space | R/pharmacometric packages [4] | Experimental design optimization |
| Collinearity Analysis | Both | Examination of parameter correlations | Custom implementation [16] | Diagnosing correlation-based non-identifiability |
| Sensitivity Matrix | Practical | Analysis of output sensitivity to parameters | R/pharmacometric packages [4] | Identifying insensitive parameters |
Table 2: Common Parameter Correlations in Inflammation Models
| Correlation Type | Typical Manifestation | Impact on Model | Resolution Strategies |
|---|---|---|---|
| Product Correlation | Parameters appearing only as products (e.g., β×π in viral replication models) [14] | Individual parameters cannot be uniquely identified | Rewrite model using composite parameters |
| Sum Correlation | Parameters appearing only in summation | Relative contributions cannot be distinguished | Incorporate prior information on parameter ratios |
| Input-Output Equivalence | Different mechanisms producing identical outputs | Model structure ambiguity | Add intermediate measurements |
| Time-Scale Correlation | Parameters affecting same temporal dynamics | Individual rate constants unidentifiable | Design experiments with multiple time resolutions |
Experimental Protocols for Identifiability Assessment
Protocol 1: Profile Likelihood Analysis for Practical Identifiability
Purpose: To assess practical identifiability of parameters given experimental data.
Materials: Dataset of time-course measurements (e.g., cytokine concentrations, viral titers), mathematical model implemented in suitable software (MATLAB, R, or Python), optimization algorithm.
Procedure:
- Estimate maximum likelihood parameters by fitting your model to the experimental data
- Select a parameter of interest (θ_i) and define a range of values around its optimum
- For each fixed value of θ_i in this range, re-optimize all other parameters to maximize likelihood
- Plot the optimized likelihood values against the fixed θ_i values
- Analyze the shape of the likelihood profile: sharply peaked profiles indicate identifiable parameters, while flat profiles suggest practical non-identifiability [16] [17]
Interpretation: The likelihood profile reveals whether the data contains sufficient information to uniquely estimate each parameter. Flat profiles indicate that the parameter cannot be constrained by the available data.
Protocol 2: Structural Identifiability Analysis with Differential Algebra
Purpose: To determine whether model parameters can be uniquely identified from perfect, noise-free data.
Materials: ODE model of inflammation, StructuralIdentifiability.jl package [18].
Procedure:
- Install StructuralIdentifiability.jl package in Julia
- Define your ODE model, specifying states, parameters, inputs, and observed outputs
- Run the
assess_identifiabilityfunction on your model - Interpret the output, which classifies each parameter as globally identifiable, locally identifiable, or unidentifiable
- For unidentifiable parameters, the software may provide relationships between parameters
Interpretation: Structurally unidentifiable parameters cannot be uniquely estimated even with perfect data, indicating fundamental issues with model structure or observation scheme [18] [4].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Computational Tools for Identifiability Analysis
| Tool/Software | Primary Function | Application in Inflammation Models | Implementation Considerations |
|---|---|---|---|
| StructuralIdentifiability.jl | Structural identifiability analysis | Analyzing ODE models of cytokine networks [18] | Requires Julia programming knowledge |
| DAISY Software | Structural identifiability via differential algebra | Examining within-host pathogen dynamics models [20] [4] | Handles rational ODE systems |
| Profile Likelihood Methods | Practical identifiability assessment | Determining parameter estimability from noisy data [16] [17] | Can be implemented in multiple environments |
| Markov Chain Monte Carlo | Bayesian parameter estimation | Characterizing parameter uncertainties in complex models [15] | Computationally intensive for large models |
| Sensitivity Analysis Tools | Identifying influential parameters | Prioritizing parameters for estimation [4] | Helps focus on identifiable parameters |
| Piprofurol | Piprofurol, CAS:40680-87-3, MF:C26H33NO6, MW:455.5 g/mol | Chemical Reagent | Bench Chemicals |
| Ppack dihydrochloride | Ppack dihydrochloride, CAS:82188-90-7, MF:C21H33Cl3N6O3, MW:523.9 g/mol | Chemical Reagent | Bench Chemicals |
Methodological Workflows
Diagram 1: Identifiability Assessment Workflow - This diagram illustrates the integrated process for evaluating both structural and practical identifiability in mathematical models of inflammation.
Diagram 2: Resolution Strategies for Non-Identifiability - This decision framework outlines pathways for addressing different types of non-identifiability in inflammation models.
Troubleshooting Guides & FAQs
FAQ 1: What are the most common sources of identifiability issues when fitting within-host viral dynamics models to data?
Identifiability issues commonly arise from two main sources: the model's inherent structure and practical limitations of the available data [14].
- Structural Non-Identifiability: This occurs when the model's structure makes it theoretically impossible to uniquely estimate parameters, even with perfect, noise-free data. A common cause is parameter correlation, where changes in one parameter can be perfectly compensated for by changes in another, leading to the same model output [14]. For example, in basic viral dynamics models, parameters for infection rate and viral production can be correlated.
- Practical Non-Identifiability: This occurs when the model is structurally identifiable, but the available data is too scarce, noisy, or lacks sufficient information to reliably estimate the parameters [14] [21]. For instance, fitting a complex model with many parameters to only viral titer data, without immune cell counts, often leads to practical identifiability problems [14].
FAQ 2: My model is structurally identifiable, but parameter estimates have wide confidence intervals. How can I improve practical identifiability?
If your model is structurally identifiable but parameters are not practically identifiable, consider these strategies:
- Increase Data Frequency and Diversity: Collect data at more time points, especially during dynamic phases like the initial peak and decline of viral load. Incorporate data for additional model variables; for example, using both daily virus titers and adaptive immune cell data significantly improves the practical identifiability of parameters in influenza models [14].
- Incorporate Prior Knowledge: Use Bayesian estimation methods, which allow you to incorporate prior distributions for parameters based on previous studies or biological knowledge. This can constrain the parameter space and improve identifiability [22].
- Optimal Experimental Design: Employ computational frameworks to design experiments that maximize the information gain for parameter estimation. This involves identifying critical time points for data collection that make all model parameters practically identifiable [21].
- Model Reduction: If certain parameters remain non-identifiable, consider whether the model can be simplified by fixing well-known parameters or by using a less complex model structure that still captures the essential biology [14].
FAQ 3: How can I check for identifiability in my mathematical model before conducting expensive experiments?
A rigorous model validation pipeline should be followed before parameter estimation [14] [21]:
- Structural Identifiability Analysis: Use dedicated software tools to check if your model is theoretically identifiable. Tools like
StructuralIdentifiability.jl(Julia) orSIAN(MATLAB) can perform this analysis using differential algebra or other methods [22] [14]. - Practical Identifiability Analysis: After confirming structural identifiability, assess practical identifiability using the available or planned data. Profile likelihood or Fisher Information Matrix (FIM) analysis can be used. A novel framework proves that practical identifiability is equivalent to the invertibility of the FIM [21]. Eigenvalue decomposition of the FIM can pinpoint which specific parameter combinations are non-identifiable [21].
The table below summarizes a typical analysis workflow and its outcomes for different types of within-host models.
Table 1: Identifiability Analysis of Example Within-Host Models
| Model Name | Key Features | Data Used for Fitting | Typical Identifiability Findings |
|---|---|---|---|
| Basic Target Cell Model | Target cells (T), infected cells (I), virus (V) [14] | Viral titer data | Often structurally identifiable but may suffer from practical non-identifiability due to parameter correlations [14]. |
| Model with Eclipse Phase | Adds eclipse phase (Iâ‚) before productive infection (Iâ‚‚) [14] | Viral titer data | Improved ability to capture delays; however, some parameters related to infected cell loss may still be non-identifiable with virus data alone [14]. |
| Model with Adaptive Immunity | Adds effector CD8+ T cells (E) explicitly [14] | Viral titer + immune cell data | Significantly improved practical identifiability of parameters related to viral clearance and infected cell death when both data types are used [14]. |
| LCMV-CTL Response Model | Models acute LCMV infection with Cytotoxic T Lymphocytes (CTL) [22] | Viral load and CTL kinetics data | Structural identifiability depends on observability and initial conditions. Bayesian approach can estimate posterior distributions, revealing that bilinear terms may need refinement [22]. |
Experimental Protocols
To address identifiability challenges, the design of experiments and model calibration must be meticulous. The following protocol outlines a robust methodology.
Protocol: A Framework for Model Identifiability Analysis and Refinement
Objective: To systematically diagnose and resolve identifiability issues in mathematical models of acute viral infection (e.g., LCMV).
Materials:
- Mathematical model (ODE/PDE system)
- Experimental dataset (e.g., viral titers, immune cell counts)
- Computational software for identifiability analysis (e.g.,
StructuralIdentifiability.jl,SIAN) and parameter estimation (e.g.,DynamicHMC.jlfor Bayesian estimation [22])
Workflow Diagram:
Methodology:
Structural Identifiability Check:
- Use differential algebraic methods (e.g., via
StructuralIdentifiability.jlpackage) to verify that all model parameters are globally or locally identifiable from the perfect, noise-free model output [22]. - Troubleshooting: If the model is structurally non-identifiable, find the source (e.g., parameter correlations) and propose additional assumptions. This may involve fixing a well-known parameter, simplifying the model, or re-parameterizing it [14].
- Use differential algebraic methods (e.g., via
Practical Identifiability Assessment:
- Using the actual (noisy) experimental data, perform a practical identifiability analysis. A reliable method is to compute the Fisher Information Matrix (FIM). According to recent frameworks, the invertibility of the FIM is a necessary and sufficient condition for all parameters to be practically identifiable [21].
- Perform eigenvalue decomposition (EVD) on the FIM. Eigenvalues equal to zero indicate that the corresponding parameter combinations (eigenvectors) are practically non-identifiable [21].
Addressing Non-Identifiability:
- Optimal Experimental Design: If parameters are not practically identifiable, use an algorithm to find time points for data collection that maximize the information content. The goal is to design an experiment that results in an invertible FIM [21].
- Parameter Regularization: For non-identifiable parameters, incorporate regularization terms based on the eigenvectors from the FIM's EVD. This technique helps constrain the parameter space during fitting, making all parameters practically identifiable [21].
- Bayesian Estimation: Implement a Bayesian approach (e.g., using Hamiltonian Monte Carlo in
DynamicHMC.jl) to estimate posterior distributions for parameters. This explicitly handles uncertainty and incorporates prior knowledge, which can mitigate identifiability problems [22].
Model Refinement:
- The identifiability analysis may suggest model structural flaws. For example, in LCMV models, Bayesian estimation of posterior distributions suggested that a bilinear term for virus-CTL interaction was inadequate and should be refined to a bounded-rate (e.g., Michaelis–Menten) formulation [22].
Research Reagent Solutions
The following table lists key reagents and computational tools essential for conducting the experiments and analyses described in this case study.
Table 2: Essential Research Reagents and Tools for LCMV Modeling Studies
| Item Name | Function / Description | Application in Identifiability Research |
|---|---|---|
| LCMV (Armstrong & Clone 13) | Armstrong strain causes acute infection. Clone 13 strain establishes persistent chronic infection [23]. | Used to generate kinetic data (viral load, immune cell counts) for model calibration and to study acute vs. chronic infection dynamics [22] [23]. |
| P14 TCR-Transgenic Mice | Genetically modified mice with T cell receptors specific for the LCMV glycoprotein peptide GP33-41 [23]. | Provides a traceable population of CD8+ T cells for precise quantification of antigen-specific immune responses, improving data quality for model fitting [23]. |
| Vaccinia virus expressing OVA (VV-OVA) | A virus engineered to express Ovalbumin (OVA) antigen [23]. | Used in challenge experiments to test T cell functionality against new antigens in chronically infected hosts, informing model predictions on immune dysfunction [23]. |
Computational Tool: StructuralIdentifiability.jl |
A Julia-based software package for analyzing structural identifiability of ODE models [22]. | Used for the initial, theoretical check of whether model parameters can be uniquely identified before data collection [22]. |
Computational Tool: DynamicHMC.jl |
A Julia-based package for Bayesian parameter inference using Hamiltonian Monte Carlo [22]. | Estimates posterior distributions of parameters, quantifying uncertainty and helping to resolve practical identifiability issues through prior information [22]. |
| Bone Marrow-Derived Dendritic Cells (BMDCs) | Dendritic cells generated in vitro from bone marrow precursors using GM-CSF [23]. | Used to study antigen presentation and T cell priming capacity under different infection conditions (naive, acute, chronic), providing data for modeling immune cell interactions [23]. |
Tools and Techniques: A Practical Toolkit for Identifiability Analysis
Differential Algebra and the Laplace Transform for Structural Analysis
Troubleshooting Guides and FAQs
This technical support resource addresses common challenges researchers face when performing structural analysis on mathematical models of inflammation.
Frequently Asked Questions
Q1: My high-index Differential-Algebraic Equation (DAE) model fails during numerical simulation. What structural issue might be causing this? High-index DAEs (index > 1) often lead to numerical instability because they contain hidden constraints that are not explicitly formulated [24]. This is a common problem in models of biological systems like inflammation where conservation laws or rapid equilibria create algebraic dependencies. The dummy derivatives method is a proven technique for index reduction that can resolve this [24].
Q2: How can I determine if my model's parameters are uniquely identifiable from the available experimental data? Perform a structural identifiability analysis before parameter estimation [8]. A profile likelihood analysis can determine if parameters are locally identifiable, which is crucial for ensuring your model yields reliable, unique parameter estimates from cytokine time-series data [8].
Q3: Can the Laplace Transform handle the complex, nonlinear interactions typical of inflammatory signaling pathways? The standard Laplace Transform is most directly applicable to linear, time-invariant systems. For nonlinear model components, a common approach is to analyze the linearized system around a steady state (e.g., homeostasis or a pathological equilibrium) [25] [26]. This facilitates local stability analysis and transfer function representation.
Q4: What is the most efficient way to compute the inverse Laplace Transform for my model's output function? For complex functions where analytical inversion is difficult, numerical techniques for Laplace transform inversion are recommended [26]. These methods allow you to obtain the time-domain solution, which can be directly compared to experimental data on cytokine dynamics.
Common Error Codes and Resolutions
| Error Code / Symptom | Root Cause | Resolution Steps |
|---|---|---|
| Numerical Instability in DAE Solver | High Index (≥2) problem structure [24] | 1. Apply structural analysis to determine the index.2. Use index reduction algorithms (e.g., dummy derivatives).3. Check for consistent initial conditions. |
| Non-Unique Parameter Estimates | Structural or practical non-identifiability [8] | 1. Conduct a sensitivity analysis.2. Perform a profile likelihood analysis.3. Re-design experiments to collect more informative data. |
| Failure in Symbolic Laplace Transform | Non-rational or highly complex transfer function | 1. Check for linearity and time-invariance of the subsystem.2. Consider partial fraction decomposition.3. Use numerical inversion methods as an alternative [26]. |
| Nebicapone | Nebicapone, CAS:274925-86-9, MF:C14H11NO5, MW:273.24 g/mol | Chemical Reagent |
| Nedaplatin | Nedaplatin, CAS:95734-82-0, MF:C2H8N2O3Pt, MW:303.18 g/mol | Chemical Reagent |
Experimental Protocols for Structural Analysis
Protocol 1: Model Identifiability Analysis via Profile Likelihood
This methodology determines if a model's parameters can be uniquely identified from a given set of experimental data [8].
Primary Objective: To establish the practical identifiability of parameters in a mechanistic model of inflammation (e.g., a model featuring TNF, IL-6, and IL-10 dynamics).
Materials and Reagents:
- In silico model implemented in a suitable computational environment (e.g., MATLAB, Python).
- Experimental or synthetic time-course data for model outputs (e.g., cytokine concentrations).
Procedure:
- Parameter Estimation: For a parameter of interest (θ), find its maximum likelihood estimate (MLE), θ*.
- Profiling: Define a series of values for θ around θ*. For each fixed value of θ, re-optimize all other model parameters to minimize the goodness-of-fit measure.
- Threshold Analysis: Plot the optimized goodness-of-fit against the fixed values of θ. A uniquely identifiable parameter will show a well-defined minimum.
- Iteration: Repeat the profiling process for all unknown parameters in the model.
Protocol 2: DAE Index Reduction using the Dummy Derivatives Method
This protocol outlines the steps to reduce the index of a high-index DAE system to an index-1 problem or an ODE, making it solvable with standard numerical integrators [24].
Primary Objective: To convert a high-index DAE model into a numerically solvable form without altering the system's inherent dynamics.
Materials and Reagents:
- The high-index DAE system definition.
- Computer algebra system (e.g., Mathematica, Maple, or symbolic toolboxes).
Procedure:
- Structural Analysis: Analyze the system equations to identify the key constraints and their derivatives.
- Differentiate Constraints: Select and differentiate the algebraic constraints that are necessary to reduce the index.
- Introduce Dummy Derivatives: Define new variables (the "dummy derivatives") to represent the time derivatives of the algebraic variables.
- System Replacement: Replace the original algebraic equations with their differentiated forms and append the definitions of the dummy derivatives to the system.
- Consistency Check: Ensure that the initial conditions for the new, larger system are consistent with the original constraints.
Model Analysis Workflows
The following diagram illustrates the logical workflow for analyzing a model's structure and identifiability before proceeding with simulation and parameter fitting.
The Scientist's Toolkit: Research Reagent Solutions
This table details key computational and mathematical "reagents" essential for the structural analysis of models in inflammation research.
| Item Name | Function / Purpose | Example Application in Inflammation |
|---|---|---|
| Dummy Derivatives Method | A systematic algorithm for reducing the index of a high-index DAE system [24]. | Enables stable simulation of complex cytokine networks with fast equilibrium steps. |
| Laplace Transform | Converts linear differential equations into algebraic equations in the s-domain, simplifying solution finding and analysis of system structure [25] [26]. | Analyzing the input-output response (e.g., LPS stimulus to TNF output) of a linearized inflammation subsystem. |
| Profile Likelihood Analysis | A statistical method for assessing practical (numerical) identifiability of model parameters [8]. | Determining if cytokine decay rate parameters can be uniquely estimated from time-course data. |
| Transfer Function | An s-domain representation (ratio of output to input) that defines the dynamic characteristics of a linear system [26]. | Quantifying the gain and phase shift between an inflammatory stimulus and a specific cytokine output. |
| Sensitivity Analysis | Quantifies how changes in model parameters affect model outputs [8] [9]. | Identifying which reaction rate most strongly influences peak IL-6 concentration, guiding targeted interventions. |
| Nefazodone | High-purity Nefazodone HCl for psychiatric research. A SARI compound for studying depression mechanisms. For Research Use Only. Not for human consumption. | |
| PR-619 | PR-619, CAS:2645-32-1, MF:C7H5N5S2, MW:223.3 g/mol | Chemical Reagent |
Frequently Asked Questions (FAQs)
1. What is StructuralIdentifiability.jl and why is it important for modeling inflammatory diseases?
StructuralIdentifiability.jl is a Julia package that determines whether parameters in mathematical models can be uniquely identified from ideal, noise-free data. For inflammation research, this is a crucial prerequisite before estimating parameters from experimental measurements. It ensures that your model's parameters for immune response dynamics, cytokine production, or complement activation are theoretically determinable, preventing unreliable conclusions from clinical or experimental data [27] [4] [28].
2. My model involves non-integer exponents, which is common in phenomenological growth models of disease. Can I analyze it with this package?
Yes, but it requires reformulation. You can introduce additional state variables to handle non-integer power exponents, making the model compatible with the differential algebra methods used by StructuralIdentifiability.jl. This approach has been successfully applied to models like the Generalized Growth Model (GGM) and Generalized Richards Model (GRM) in epidemiology [7].
3. What is the difference between the assess_identifiability and assess_local_identifiability functions?
assess_identifiability: Checks for global identifiability. If a parameter is globally identifiable, its true value can be uniquely determined from the data.assess_local_identifiability: Checks for local identifiability. A locally identifiable parameter's value can be determined down to a finite number of possibilities within a local region [28]. For model development, it is recommended to check global identifiability first.
4. The analysis indicates some parameters are unidentifiable. What are my options? When parameters are unidentifiable, you can:
- Find Identifiable Combinations: Use the
find_identifiable_functionsfunction. This will find combinations of parameters (e.g., sums or products) that are identifiable, even if individual parameters are not [27]. - Incorporate Prior Knowledge: Use the
known_pargument to specify any parameters whose values are already known from previous literature or experiments. This can make other parameters identifiable [28]. - Adjust Measured Quantities: If possible, change or add to the model outputs you plan to measure experimentally.
5. How reliable are the results from this package?
The algorithms used are randomized but provide a very high probability of correctness. By default, the probability threshold is set to 99%. You can increase this bound (e.g., to 99.9%) using the prob_threshold argument, making the results even more conservative [27] [28].
Troubleshooting Common Issues
Problem: Installation fails or the package does not precompile correctly.
- Solution: Ensure you are using a compatible version of Julia. The package is regularly updated, so check the GitHub repository for the latest version requirements. Start a fresh Julia session and try:
Problem: The analysis is taking too long or running out of memory for a large ODE model.
- Solution: The computational complexity depends on the model structure.
- Simplify the model: Consider if all components are essential for your research question. Reduced-order modeling has been successfully used in complex immune system models [29].
- Check local identifiability first: Use
assess_local_identifiability, which is often less computationally expensive than the global analysis [27]. - Use the
funcs_to_checkargument: Instead of analyzing all parameters, you can specify a subset of critical parameters to check, which speeds up the process [28].
Problem: The package reports that key parameters in my inflammation model are unidentifiable.
- Solution: This is a fundamental mathematical issue, not a software bug. Follow this diagnostic workflow:
- Verify observed quantities: Confirm that the
measured_quantitiesyou have defined correspond to biologically plausible measurements (e.g., serum cytokine levels, viral load). - Check for over-parameterization: Does your model have more parameters than the data can support? Compare your model's complexity to validated models of immune responses to SARS-CoV-2 or sepsis [30] [31].
- Reformulate the model: Unidentifiable parameters may indicate a structural flaw. Consult the literature; for example, identifiability analysis has been used to refine COVID-19 models [30] [32].
- Verify observed quantities: Confirm that the
Problem: I receive an error about "non-rational function" when defining my model.
- Solution: The underlying differential algebra methods require models defined by rational functions (ratios of polynomials). If your model contains non-rational terms (e.g.,
sqrt,exp), you may need to reformulate it or use an approximation. The reformulation strategy used for non-integer exponents in growth models can serve as inspiration [7].
Essential Workflow and Protocols
Basic Protocol for Structural Identifiability Analysis
- Model Definition: Define your ODE model using the
@ODEmodelmacro or create aReactionSysteminCatalyst.jl[27] [28]. - Specify Observed Quantities: Declare which model variables or combinations of variables correspond to measurable outputs using the
measured_quantitiesargument. In inflammation models, these could be viral load, interleukin concentrations (e.g., IL-6), or T cell counts [30]. - Declare Known Parameters: Use the
known_pargument to specify any parameters with values fixed from prior knowledge. - Run Identifiability Analysis: Execute
assess_identifiabilityorassess_local_identifiabilityon your model. - Interpret Results: The output is a dictionary classifying each parameter and state as
:globallyidentifiable,:locallyidentifiable, or:nonidentifiable.
The following workflow diagram visualizes the key steps and decision points in this process.
Protocol for Integrating Analysis with Inflammation Research
This protocol connects identifiability analysis directly with experimental research on inflammation, providing a methodology to ensure model parameters for immune responses can be determined from typical experimental measurements [30] [31].
- Define the Pathophysiological Context: Clearly state the inflammatory condition being modeled (e.g., sepsis, SARS-CoV-2 infection, acute pancreatitis).
- Map Biology to Mathematics: Formulate the ODE model, incorporating key immune components (e.g., innate/adaptive immunity, specific cytokines, tissue damage).
- Align with Experimental Data: Define
measured_quantitiesbased on available or planned experimental data (e.g., viral load from PCR, IL-6 from serum ELISA, immune cell counts from flow cytometry). - Incorporate Prior Knowledge: Use
known_pto fix parameter values obtained from published literature or previous experiments. - Execute Identifiability Analysis: Run the analysis using the basic protocol.
- Iterate and Validate: If unidentifiable parameters are found, refine the model or measurement strategy. Validate the final identifiable model against unseen experimental data [30] [29].
Research Reagent Solutions: Computational Tools for Identifiability
The following table details key software and computational "reagents" essential for performing robust identifiability analysis in mathematical immunology.
| Item/Software | Primary Function | Relevance to Inflammation Modeling |
|---|---|---|
StructuralIdentifiability.jl |
Core engine for determining structural identifiability of ODE model parameters. | Foundational for ensuring immune response model parameters (e.g., viral replication rates, immune cell activation) are theoretically measurable from data [27] [32]. |
Catalyst.jl |
A Julia package for modeling and simulating chemical reaction networks. | Provides a high-level interface to define reaction network models of biochemical inflammation pathways (e.g., complement system, cytokine signaling) and seamlessly connect them to StructuralIdentifiability.jl [28]. |
| BioGears Physiology Engine | A whole-body, open-source mathematical model of human physiology. | Serves as a platform for building and testing complex, multi-compartment models of systemic inflammatory conditions like sepsis, linking immune dynamics to clinical physiology [31]. |
| BioUML Platform | An open-source platform for systems biology and kinetic modeling. | Used in developing and calibrating modular immune response models, such as for COVID-19, facilitating model reuse and integration into larger frameworks like a Digital Twin [30]. |
| Differential Algebra Method | The underlying mathematical methodology used by StructuralIdentifiability.jl. |
Provides the theoretical foundation for eliminating unobserved state variables (e.g., internal cellular states) to determine parameter identifiability from observable outputs (e.g., blood cytokine levels) [4] [7]. |
Data Presentation: Key Identifiability Analysis Outputs
The output of an identifiability analysis is a classification of model quantities. The table below summarizes the possible results and their implications for your research.
| Classification | Meaning | Implication for Research |
|---|---|---|
| Globally Identifiable | The parameter's value can be uniquely determined from the perfect data. | Ideal outcome. You can proceed with parameter estimation from experimental data with high confidence. |
| Locally Identifiable | The parameter's value can be determined, but only down to a finite number of possibilities in a local region. | Acceptable outcome. Parameter estimation is possible but may be sensitive to initial guesses for the optimizer. |
| Nonidentifiable | The parameter's value cannot be determined from the data; infinitely many values can produce the same model output. | Problematic outcome. The model or data collection strategy must be revised (e.g., by finding identifiable combinations or measuring additional quantities) before reliable parameter estimation is possible [27] [28]. |
Advanced Configuration and Optimization
Handling Complex Observations: The measured_quantities argument can accept algebraic expressions, not just single variables. This is useful if your experimental assay measures a sum of species (e.g., total versus phosphorylated protein) [28].
Targeted Analysis for Large Models: For large models, use the funcs_to_check argument to analyze only a specific subset of parameters or custom expressions, significantly reducing computation time [28].
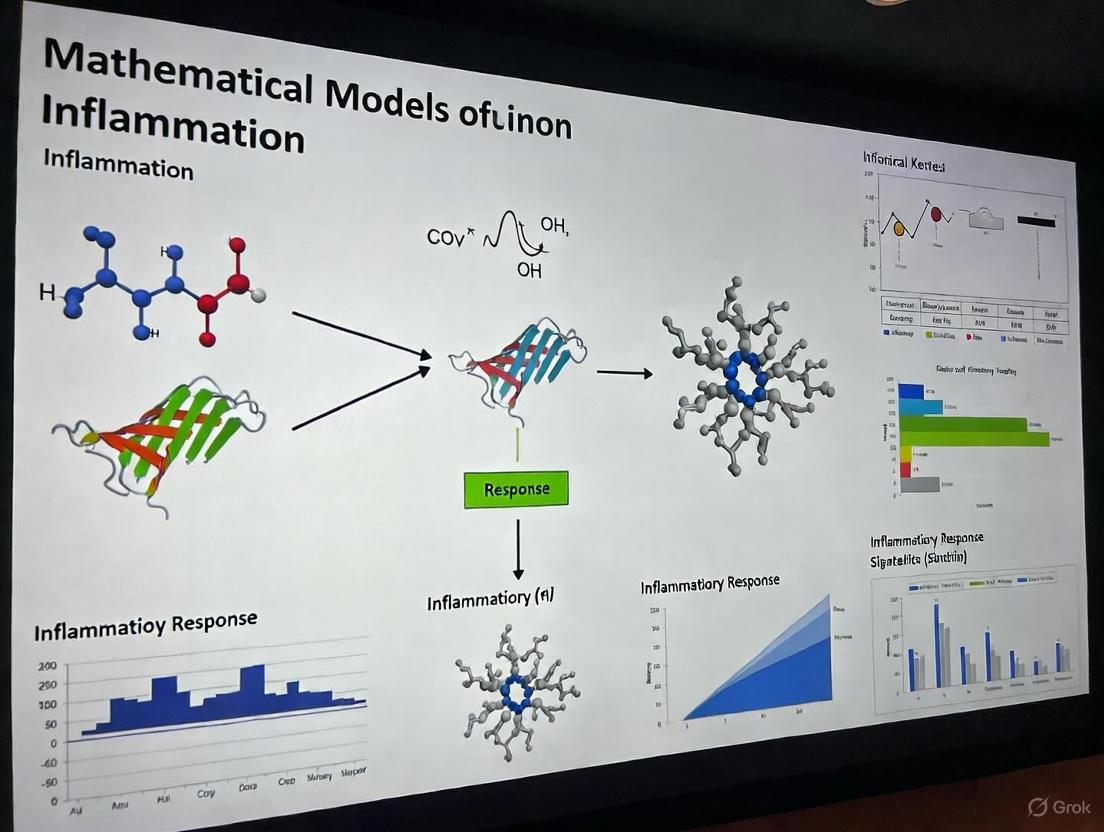
Global Sensitivity Analysis with RS-HDMR for High-Dimensional Models
Mathematical models of acute inflammation are crucial for understanding the complex dynamics of immune responses to infection and injury. These models typically consist of numerous coupled ordinary differential equations (ODEs) with many uncertain parameters [33]. A significant challenge in this field is parameter identifiability - the difficulty in determining unique parameter values that generate model outputs consistent with experimental data. Global Sensitivity Analysis (GSA) using Random Sampling-High Dimensional Model Representation (RS-HDMR) has emerged as a powerful approach to address these identifiability issues by systematically quantifying how parametric uncertainties affect model outputs [33] [34].
The complex, non-linear nature of inflammation has made it difficult to directly translate results from animal studies to clinical trials [33]. Traditional local sensitivity methods, which vary one parameter at a time while keeping others constant, are insufficient for capturing the complex parameter interactions characteristic of biological systems. Variance-based global methods like RS-HDMR provide a more comprehensive approach by exploring the entire parameter space simultaneously, making them particularly suitable for high-dimensional models of inflammatory processes [33] [35].
Core RS-HDMR Methodology
Theoretical Foundation
RS-HDMR is a metamodeling technique that approximates the input-output relationship of complex models by decomposing the model output variance into contributions from individual parameters and their interactions [35]. The fundamental HDMR equation represents the model output ( f(x) = f(x1, \ldots, xn) ) as:
[ f(x) = f0 + \sum{i=1}^n fi(xi) + \sum{1 \leq i < j \leq n} f{ij}(xi, xj) + \cdots + f{12 \ldots n}(x1, x2, \ldots, xn) ]
where:
- ( f_0 ) denotes the zeroth-order mean effect
- ( fi(xi) ) represents the first-order contribution of parameter ( x_i )
- ( f{ij}(xi, xj) ) captures the second-order interaction effects between parameters ( xi ) and ( x_j )
- Higher-order terms represent increasingly complex interactions [35]
For most practical applications, including inflammation models, the expansion can be truncated at the second order while maintaining sufficient accuracy, significantly reducing computational complexity [35].
RS-HDMR Algorithm Workflow
Table: Key Steps in RS-HDMR Implementation
| Step | Description | Considerations for Inflammation Models |
|---|---|---|
| Parameter Space Definition | Establish bounds for all model parameters based on biological knowledge | Use wide bounds to account for parametric uncertainty in biological systems |
| Sample Generation | Create input parameter samples using Monte Carlo or quasi-random sequences (Sobol' sequence recommended) | 10³-10ⴠsamples typically sufficient for models with 50+ parameters [33] |
| Model Evaluation | Run the model for each parameter set to generate output data | Parallel computing essential for computationally expensive models |
| Metamodel Construction | Build approximate functions using orthonormal polynomials | Second-order expansion often captures >95% of output variance [35] |
| Sensitivity Index Calculation | Compute variance-based Sobol' indices from component functions | Identify parameters driving uncertainty in inflammatory damage metrics |
Troubleshooting Common RS-HDMR Implementation Issues
Insufficient Metamodel Accuracy
Problem: The RS-HDMR metamodel explains less than 90% of output variance, indicating poor approximation of the original model.
Solutions:
- Increase sample size by 25-50% and reassess metamodel performance
- Check parameter bounds for biological plausibility; overly wide bounds can reduce metamodel accuracy
- Consider increasing the expansion order to third-order terms for highly nonlinear systems
- Implement the D-MORPH-HDMR extension, which improves accuracy with limited samples by solving linear algebraic equations with constrained solutions [36]
Verification Protocol:
- Generate a new validation set of 100-200 parameter samples
- Compare outputs from original model and RS-HDMR metamodel
- Calculate R² values for each model output of interest
- Accept metamodel if R² > 0.9 for all critical outputs [35]
Computational Resource Limitations
Problem: Model evaluation time prohibits the required number of Monte Carlo samples.
Solutions:
- Implement the Morris screening method as a preliminary step to identify and fix non-influential parameters [35]
- Use quasi-random Sobol sequences instead of random sampling for better coverage with fewer samples [35]
- Employ parallel computing frameworks to distribute model evaluations across multiple processors
- For extremely computationally intensive models, use a two-stage approach: initial screening with fewer samples followed by focused RS-HDMR on influential parameters
Interpretation of Sensitivity Indices
Problem: Confusion in ranking parameters based on first-order versus total-effect sensitivity indices.
Guidance:
- First-order indices (( S_i )) measure the individual contribution of each parameter to output variance
- Total-effect indices (( S_{Ti} )) include both individual and all interaction effects
- Parameters with large differences between ( Si ) and ( S{Ti} ) participate in significant interactions
- For identifiability analysis, focus on parameters with high first-order indices as they are independently identifiable
- Parameters with low first-order but high total-effect indices may require dedicated experiments to resolve interactions [33] [35]
RS-HDMR Application to Inflammation Models: Case Examples
Acute Inflammatory Response to LPS
In a study of acute inflammation induced by lipopolysaccharide (LPS), researchers applied RS-HDMR to a 51-parameter ODE model to identify key drivers of whole-animal damage and dysfunction [33]. The analysis revealed that inflammatory damage was highly sensitive to parameters affecting IL-6 activity during different stages of acute inflammation, highlighting this cytokine as a critical control point [33].
Table: Key Sensitive Parameters in LPS-Induced Inflammation Model
| Parameter Category | Sensitivity Ranking | Biological Process | Identifiability Priority |
|---|---|---|---|
| IL-6 production and clearance | High | Pro-/anti-inflammatory balance | Critical |
| Nitric Oxide (NO) synthesis | Medium-High | Anti-inflammatory response | High |
| TNF-α dynamics | Medium | Early pro-inflammatory signaling | Medium |
| IL-10 regulation | Medium | Anti-inflammatory feedback | Medium |
| Neutrophil recruitment | Low-Medium | Innate immune response | Low |
The RS-HDMR analysis further revealed bimodal behavior in the system, where the Area Under the Curve for IL-6 (AUCIL6) showed two distinct peaks representing healthy response and sustained inflammation [33]. This finding demonstrated how RS-HDMR can identify critical transitions in inflammatory outcomes.
Genetic Circuit Optimization
While not specific to inflammation, a study optimizing genetic circuits demonstrated RS-HDMR's capability to guide biological engineering [34]. The method correctly identified that inverter output was more sensitive to mutations in the ribosome-binding site upstream of the cI coding region than mutations in the OR1 region of the PR promoter [34]. This approach can be adapted for identifying optimal intervention targets in inflammatory signaling networks.
Frequently Asked Questions (FAQs)
Q1: What sample size is needed for reliable RS-HDMR analysis of inflammation models? For typical inflammation models with 50-100 parameters, 1000-5000 samples generally provide sufficient accuracy. Start with 1000 samples and increase until metamodel R² > 0.9. The exact requirement depends on the degree of nonlinearity and parameter interactions in your specific model [35].
Q2: How does RS-HDMR compare to other GSA methods for identifiability analysis? RS-HDMR provides several advantages: (1) It requires only one set of Monte Carlo samples, unlike traditional Sobol' method which needs multiple specialized samples; (2) It simultaneously generates a accurate metamodel for further analysis; (3) It efficiently handles high-dimensional spaces with parameter interactions [35]. For inflammation models specifically, RS-HDMR has successfully identified key regulatory parameters missed by local methods [33].
Q3: Can RS-HDMR handle correlated parameters common in biological systems? Standard RS-HDMR assumes parameter independence. For correlated parameters, extended methods like covariance-based HDMR are available [35]. In practice, for mild correlations, the standard method often remains effective, but strong correlations should be addressed through model reparameterization or using specialized correlation-handling extensions.
Q4: What software tools are available for implementing RS-HDMR? The GUI-HDMR software package (MATLAB-based) provides a user-friendly implementation with graphical interface [35]. For programming-based approaches, Python and R implementations are available through various scientific computing libraries. Custom implementation is also feasible based on the mathematical framework described in the literature [35].
Q5: How can RS-HDMR results guide experimental design in inflammation research? RS-HDMR sensitivity indices directly identify which biochemical parameters most influence model outputs. This allows prioritization of measurement efforts for parameters with high sensitivity indices, significantly improving model identifiability. For example, in inflammation models, IL-6-related parameters often show high sensitivity, guiding targeted experiments to better quantify IL-6 dynamics [33].
Essential Research Reagents and Computational Tools
Table: Key Resources for RS-HDMR in Inflammation Research
| Resource Category | Specific Tools/Reagents | Application Purpose | Implementation Notes |
|---|---|---|---|
| Software Tools | GUI-HDMR (MATLAB) | User-friendly RS-HDMR implementation | Ideal for researchers with limited programming experience [35] |
| D-MORPH-HDMR extension | Enhanced accuracy with limited samples | Particularly useful for computationally expensive models [36] | |
| Custom Python/R scripts | Flexible implementation for specific needs | Requires programming expertise but offers maximum flexibility | |
| Experimental Reagents | LPS preparations | Inducing inflammatory response in experimental models | Enables model calibration and validation [33] [8] |
| Cytokine measurement assays | Quantifying TNF-α, IL-6, IL-10 dynamics | Critical for parameter estimation in inflammation models [33] | |
| Immune cell isolation kits | Studying specific cell population dynamics | Enables cell-specific parameter estimation |
Workflow Visualization
RS-HDMR Identifiability Enhancement Workflow
Advanced Applications and Future Directions
Recent advances in RS-HDMR methodology have expanded its applications in inflammation research. The integration of information-theoretic approaches with mathematical modeling shows promise for deciphering causal relationships in inflammatory networks, such as connections between arachidonic acid metabolism and cytokine secretion [37]. Additionally, the development of multi-scale inflammation models that incorporate cellular, tissue, and whole-organism levels presents new challenges and opportunities for RS-HDMR application [8].
Future methodological developments should focus on:
- Enhanced computational efficiency for large-scale models
- Improved handling of parameter correlations common in biological systems
- Integration with machine learning approaches for pattern recognition in sensitivity results
- Application to personalized medicine through patient-specific parameterization
As mathematical models of inflammation continue to increase in complexity, RS-HDMR and related global sensitivity analysis methods will remain essential tools for addressing fundamental identifiability challenges and translating computational insights into biological understanding and therapeutic applications.
Bayesian Approaches and Hamiltonian Monte Carlo with DynamicHMC.jl
Troubleshooting Guides
Failed to Find Valid Initial Parameters
Problem: Your sampler fails with an error stating it failed to find valid initial parameters in {N} tries [38].
Explanation: This error occurs when the Hamiltonian Monte Carlo (HMC) sampler cannot locate a starting position where both the log probability density and its gradient are finite and not NaN [38]. In the context of inflammation models, this often happens when parameters fall outside biologically plausible ranges.
Common Causes and Solutions:
NaNGradients: Often caused by invalid parameter values in distributions or functions [38].- Diagnosis: Manually evaluate your model's gradient at different parameter values.
- Solution: Check for problematic constructions like
truncated(Normal(0,1), Inf, Inf). Remove unnecessary bounds or useBijectors.jlfor constrained parameters [38].
-InfLog Density: Occurs when initial parameters place the model in an impossible biological state [38].- Example: In inflammation models, this might happen if a cytokine concentration is initialized as negative.
- Solution: Override the default initialization strategy. Instead of
InitFromUniform(-2, 2), specify biologically plausible initial ranges [38]:
HMC Gets Stuck at Tiny Local Maxima
Problem: Your chains appear to converge to different regions of parameter space in different runs, even though the posterior appears unimodal [39].
Explanation: This behavior, where "HMC saw the posterior as multimodal" despite visual evidence to the contrary, can occur due to several factors [39]:
Solutions:
Reparameterization: Transform parameters to make the posterior geometry more friendly for HMC [39] [40].
- For concentration parameters bounded at
[0, ∞), use a logarithmic transform. - For proportions bounded at
[0, 1], use a logit transform.
- For concentration parameters bounded at
Jittering: Add random noise to the step size to help escape flat regions [40].
Curvature Adaptation: Ensure you're using sufficient warmup samples for the sampler to adapt to the local curvature of your inflammation model [40].
ForwardDiff Type Errors
Problem: You encounter MethodError: no method matching Float64(::ForwardDiff.Dual{...}) when using automatic differentiation [38].
Explanation: This occurs when your model code contains type-unstable operations that cannot handle the ForwardDiff.Dual number types used for automatic differentiation [38].
Solutions:
- Ensure Type Stability: Use
DynamicPPL.DebugUtils.model_warntypeto check for type instabilities in your model [41]. - Avoid Type Annotations: Don't force specific types like
Vector{Float64}in your model code. - Use AD-Compatible Functions: Ensure all functions in your model (including custom ones) can handle
Dualnumbers.
Frequently Asked Questions
How should I handle parameter constraints in inflammation models?
Answer: Use TransformVariables.jl in combination with TransformedLogDensities.jl for domain transformations [42]. For example:
- Cytokine concentrations (must be positive): Use
logtransform - Probability parameters (bounded [0,1]): Use
logittransform - Correlation parameters: Use appropriate constraints for valid matrices
This approach automatically handles the Jacobian corrections required for proper sampling [42].
Why is my model suddenly slow after a small change?
Answer: Small changes can significantly impact performance through [41]:
- Introducing type instability
- Switching between vectorized and scalar operations
- Breaking compiler optimizations
- Causing AD backend incompatibilities
Diagnosis: Use DynamicPPL.DebugUtils.model_warntype to check for type instability and profile your log-density function to identify bottlenecks [41].
Can I use threading within my DynamicHMC model?
Answer: Yes, but with important caveats [41]:
observestatements (likelihood): Generally safe in threaded loopsassumestatements (priors/sampling): Often crash unpredictably or produce incorrect results- AD backend compatibility: Many automatic differentiation backends don't support threading
For safe parallelism, prefer vectorized operations over explicit threading with Threads.@threads [41].
Experimental Protocols for Inflammation Models
Parameter Identifiability Analysis
Purpose: Determine which parameters in your inflammation ODE model can be uniquely identified from available data [8].
Protocol:
- Local Sensitivity Analysis: Calculate partial derivatives of model outputs with respect to parameters [8]
- Profile Likelihood Analysis: Profile out each parameter while optimizing others [8]
- Identifiability Classification:
- Structurally identifiable: Parameters that are theoretically unique
- Practically identifiable: Parameters that can be estimated with available data quality
- Unidentifiable: Parameters that cannot be uniquely determined
Implementation:
Model Calibration with Experimental Data
Purpose: Estimate identifiable parameters using combined in vitro and in vivo data [8].
Protocol:
- Parameter Subset Selection: Based on sensitivity analysis, select parameters for estimation [8]
- Multi-Stage Calibration:
- First, use in vitro data to inform cellular-level parameters
- Then, use human endotoxemia data for system-level parameters [8]
- Validation: Test calibrated model against held-out data (different LPS dosages or infusion protocols) [8]
Research Reagent Solutions
Table: Essential Computational Tools for Bayesian Inflammation Modeling
| Tool/Reagent | Function | Application Context |
|---|---|---|
| DynamicHMC.jl [43] | No-U-Turn Sampler implementation | Robust posterior sampling for complex models |
| TransformVariables.jl [42] | Domain transformation with Jacobian correction | Handling biologically constrained parameters |
| LogDensityProblems.jl [42] | Standard interface for log-posteriors | Creating compatible target distributions |
| ForwardDiff.jl [38] | Automatic differentiation | Gradient calculation for HMC |
| ProfileLikelihood.jl | Parameter identifiability analysis | Determining which parameters are estimable |
| Turing.jl [38] [41] | Probabilistic programming | Alternative interface for model specification |
Workflow Visualization
Bayesian Workflow for Inflammation Models
DynamicHMC.jl Sampling Process
Inflammation Model Structure
FAQ: Core Concepts and Applications
Q1: What is a Hybrid Neural ODE (HNODE), and why is it relevant for inflammation research? A Hybrid Neural ODE (HNODE) is a modeling framework that integrates partially known mechanistic Ordinary Differential Equation (ODE) models with neural networks. The neural network acts as a universal approximator to represent unknown biological processes or unmodeled dynamics within an otherwise mechanistic system [44]. In inflammation research, where mechanisms are often only partially understood, this approach allows you to leverage established biological knowledge (e.g., core cytokine interactions) while using data to learn the missing pieces, thus creating more accurate and predictive models of the host inflammatory response [8] [44].
Q2: What are the most common identifiability issues when fitting HNODEs? The primary identifiability challenge in HNODEs is the compensation effect between the mechanistic parameters and the neural network component [44]. The flexibility of the neural network can make it difficult to uniquely determine the values of the mechanistic parameters, as different combinations of parameter values and network outputs can produce similarly accurate fits to the data. This can lead to non-identifiable parameters, where multiple values explain the data equally well, undermining the model's biological interpretability [44].
Q3: What practical steps can I take to improve parameter identifiability in my HNODE? A robust pipeline for parameter estimation and identifiability analysis involves several key steps [44]:
- Global Exploration: Treat the mechanistic parameters as hyperparameters and use global search methods like Bayesian Optimization to initially explore the parameter space.
- Model Training: Train the full HNODE model using gradient-based methods.
- Identifiability Analysis: After training, perform a practical identifiability analysis (e.g., profile likelihood) to determine which parameters are uniquely identified by the data.
- Uncertainty Quantification: For identifiable parameters, calculate confidence intervals to assess estimation uncertainty.
Troubleshooting Common Experimental Issues
Q1: My HNODE training is unstable, and the loss does not converge. What could be wrong? This is a common issue, often stemming from the combination of ODE solvers and gradient-based optimization.
- Potential Cause: Inappropriate ODE solver for "stiff" systems. Biological systems like inflammation often exhibit dynamics on vastly different timescales (e.g., rapid cytokine release vs. slow tissue repair), leading to stiff equations that cause numerical instability in non-stiff solvers [45].
- Solution: Use a stiff ODE solver, such as
Kvaerno5[45]. Additionally, implement gradient clipping (e.g., with a global norm max of 4.0) to prevent exploding gradients, a known issue in training Neural ODEs [45]. - Solution: Optimize parameters in log-space. This can help manage the large differences in parameter scales common in biological systems and stabilize the training process [45].
Q2: The neural network in my HNODE is dominating the dynamics, making the mechanistic parameters uninterpretable. How can I enforce the role of the mechanistic part? This problem is at the heart of ensuring model interpretability.
- Potential Cause: Lack of constraints on the neural network's influence, allowing it to override the mechanistic component.
- Solution: Incorporate a regularization term into the loss function that explicitly penalizes the magnitude of the neural network's output or its correlation with the mechanistic part. This encourages the model to use the neural network only for what the mechanistic model cannot explain [44].
Q3: I have limited and noisy experimental data. Will the HNODE approach still work? Yes, but it requires careful setup. The "over-parameterization" of HNODEs via the neural network makes them susceptible to overfitting on small datasets.
- Solution: Use a tailored loss function. Instead of a standard Mean Squared Error, use a mean-centered relative error. This prevents the loss from being dominated by variables with large absolute concentrations and ensures all system components are learned effectively, even with sparse data [45].
- Solution: Rigorously split your data into training and validation sets. Use the validation set to monitor for overfitting and to guide hyperparameter tuning and model selection [44].
Experimental Protocols & Workflows
A Protocol for Building an HNODE for an Inflammatory Response Model
This protocol outlines the process of constructing an HNODE to model cytokine dynamics, where some signaling pathways are unknown.
1. Problem Formulation and Mechanistic Scaffold:
- Define the System: Identify the key state variables (e.g., concentrations of TNF, IL-6, IL-10, damaged tissue) [8].
- Establish the Known Mechanics: Write down the ODEs for processes you are confident about. For example, you might know that activated immune cells produce pro-inflammatory cytokines, and that IL-10 inhibits their production [8].
d[TNF]/dt = (Production from cells) - (Decay rate) * [TNF] - Identify the Knowledge Gap: Pinpoint a process that is poorly characterized. For instance, the precise feedback mechanism controlling IL-10 expression might be unknown.
2. HNODE Integration:
- Embed the Neural Network: Replace the unknown process with a neural network. The HNODE system then becomes:
Here,
f_mechanisticcontains the known kinetics, whileNNlearns the unknown interactions from data [44].
3. Data Preparation and Preprocessing:
- Gather Data: Use experimental time-series data of the state variables (e.g., from LPS-exposure studies in humans or in vitro immune cell cultures) [8].
- Split Data: Partition data into training and validation sets across different time points [44].
- Scale Data: Normalize the data, for example by the mean observed concentration, to ensure balanced contributions to the loss function [45].
4. Model Training and Identifiability Analysis:
- Hyperparameter Tuning: Use Bayesian Optimization to perform a global search over both model hyperparameters (e.g., learning rate) and the mechanistic parameters (
θ) [44]. - Gradient-Based Training: Train the model using a gradient-based optimizer like Adam or AdaBelief, with gradients computed via the adjoint sensitivity method for efficiency [45] [44].
- Identifiability Check: Conduct a practical identifiability analysis (e.g., profile likelihood) on the trained model to determine which mechanistic parameters are uniquely identifiable given the data and the HNODE structure [44].
The following workflow diagram illustrates the complete pipeline from problem definition to identifiability analysis.
Quantitative Data from Representative Studies
The table below summarizes key quantitative findings from studies that utilize related neural ODE and hybrid approaches, providing benchmarks for expected performance.
Table 1: Performance Benchmarks in Neural ODE and Hybrid Modeling Studies
| Study / Model | Application Context | Key Performance Metric | Result | Reference |
|---|---|---|---|---|
| cd-PINN (Continuous Dependence PINN) | Solving ODEs (Logistic model, Lotka-Volterra) | Generalization Accuracy (Relative Error) | 1-3 orders of magnitude improvement over vanilla PINN | [46] |
| Low-Dimensional NODE | Pharmacokinetics (PK) Modeling | Ability to simulate new subjects | Successfully described data and simulated within observed dosing range | [47] |
| jaxkineticmodel | Metabolic Kinetic Model Training | Robust Training Convergence | Successfully trained on a benchmark of 26 SBML models | [45] |
| HNODE Pipeline | Robust Parameter Estimation | Practical Identifiability | Enabled identifiability analysis for mechanistic parameters in partially known systems | [44] |
The Scientist's Toolkit: Research Reagent Solutions
This table lists essential computational "reagents" required to implement an HNODE framework for inflammation modeling.
Table 2: Essential Computational Tools for HNODE Research
| Tool / Reagent | Function / Purpose | Relevance to Inflammation Modeling |
|---|---|---|
| JAX/Diffrax | A high-performance numerical computing library with automatic differentiation and a suite of ODE solvers. | Enables efficient computation of gradients via the adjoint method for training and provides stiff solvers necessary for multi-scale inflammatory dynamics. [45] |
| Bayesian Optimization | A global optimization strategy for tuning hyperparameters and exploring mechanistic parameter spaces. | Crucial for the initial global search of mechanistic parameters (e.g., cytokine production/decay rates) before fine-tuning, helping to avoid local minima. [44] |
| Adjoint Sensitivity Method | A technique to compute gradients of solutions with respect to parameters by solving a second ODE backwards in time. | Makes training HNODEs computationally feasible, as it is more efficient than forward sensitivity analysis for models with many parameters. [45] [44] |
| Profile Likelihood Analysis | A practical identifiability analysis method that assesses whether parameters can be uniquely estimated from available data. | Determines the reliability of estimated parameters (e.g., rate constants in your cytokine model), which is critical for biological interpretation and hypothesis generation. [44] |
| Stiff ODE Solver (e.g., Kvaerno5) | A numerical integrator designed for systems of ODEs with widely varying timescales (stiffness). | Essential for accurately simulating the inflammatory response, which involves fast-acting cytokines and slow-acting tissue repair processes. [45] |
| Pramiconazole | Pramiconazole, CAS:219923-85-0, MF:C35H39F2N7O4, MW:659.7 g/mol | Chemical Reagent |
| Oxolamine | Oxolamine, CAS:959-14-8, MF:C14H19N3O, MW:245.32 g/mol | Chemical Reagent |
The following diagram illustrates how these computational tools interact within a typical HNODE analysis workflow for a biological system.
From Unidentifiable to Robust: Strategies for Model Repair and Refinement
Parameter Set Reduction and Model Re-parameterization
Frequently Asked Questions (FAQs)
1. What is the core difference between structural and practical identifiability? Structural identifiability is a theoretical property of your model. It asks whether model parameters can be uniquely determined assuming perfect, noise-free data collected continuously over time. If a model is structurally unidentifiable, no amount or quality of real data can make its parameters identifiable. Practical identifiability, in contrast, concerns whether parameters can be accurately estimated given the limitations of real-world data—including noise, limited sampling timepoints, and sparse measurements [4] [48].
2. My complex inflammation model is unidentifiable. What is the first step I should take? The first step is to conduct a parameter sensitivity analysis, as demonstrated in mathematical models of the inflammatory response [8]. This analysis identifies which parameters your model's output is most sensitive to. You can then focus your efforts on the most influential parameters. Following this, a parameter identifiability analysis (e.g., using the Fisher Information Matrix Method) can determine which of these sensitive parameters can be reliably estimated from your data [4].
3. How can the reparameterization trick help in training probabilistic models? In models like Variational Autoencoders (VAEs), directly sampling from a latent distribution (e.g., a Gaussian) blocks gradient flow during backpropagation because the sampling operation is non-differentiable. The reparameterization trick rewrites the sampling process as a deterministic function of the model's parameters and a fixed noise source. This allows gradients to flow through the deterministic path, enabling efficient training with stochastic gradient descent [49] [50]. For a Gaussian distribution, this means sampling via ( z = \mu + \sigma \odot \epsilon ), where ( \epsilon ) is drawn from a standard normal distribution [49].
4. When should I consider re-parameterizing my model versus reducing the number of parameters? Re-parameterization is often the preferred first step when the relationship between parameters is the source of unidentifiability (e.g., if only their product is identifiable). It aims to find a new, smaller set of identifiable parameter combinations without discarding biological meaning. Parameter set reduction, which involves fixing non-identifiable parameters to constant values, should be considered when re-parameterization is not feasible or when specific parameters have been well-established by prior experiments [8] [4].
Troubleshooting Guides
Problem 1: Unidentifiable Parameters in a Model of Inflammation
Symptoms
- Failed or poor convergence when fitting the model to experimental data.
- Large confidence intervals on estimated parameter values.
- Strong correlations between different parameter estimates.
Diagnosis and Resolution Steps
Perform a Structural Identifiability Analysis: Use a tool like DAISY (Differential Algebra for Identifiability of SYstems) to check if your model is theoretically identifiable with perfect data [4]. This step can save significant time by diagnosing fundamental flaws in the model structure.
Conduct a Practical Identifiability Analysis: If the model is structurally identifiable, use a posteriori methods to diagnose issues with your specific dataset. The Fisher Information Matrix Method (FIMM) is particularly recommended, as it can handle random effects and provides a continuous measure of identifiability [4]. Profile Likelihood Analysis is another common method used for this purpose [8].
Apply a Solution:
- Reparameterize: Find a new, lower-dimensional parameterization. For example, if parameters
k1andk2always appear as a productP = k1*k2in your model equations, estimate the combined productPinstead of the individual parameters. - Reduce the Parameter Set: Fix non-identifiable or less sensitive parameters to literature values [8]. The table below summarizes this decision process.
- Reparameterize: Find a new, lower-dimensional parameterization. For example, if parameters
| Approach | Description | Best Used When |
|---|---|---|
| Reparameterization | Transforming parameters into a new, smaller set of identifiable combinations. | Parameters are correlated, and a biologically meaningful combined parameter can be defined [49]. |
| Parameter Fixing | Manually setting non-identifiable parameters to constant values from literature. | Some parameters are well-known and can be fixed to reduce complexity [8]. |
| Sensitivity-Based Reduction | Removing parameters to which the model output is least sensitive. | Facing practical identifiability issues, and some parameters have a negligible effect on outputs [8]. |
The following workflow diagram illustrates the diagnostic process for unidentifiable parameters:
Problem 2: High Variance in Gradient Estimates During Stochastic Optimization
Symptoms
- Unstable or fluctuating loss curves during training of machine learning or variational inference models.
- Failure of the model to converge despite seemingly appropriate hyperparameters.
Diagnosis and Resolution Steps
Identify the Source: This problem is common in models with stochastic nodes, such as Variational Autoencoders (VAEs), where random sampling blocks gradient flow [49] [50].
Apply the Reparameterization Trick: Replace the non-differentiable sampling step with a differentiable function. The table below shows common reparameterizations.
| Distribution | Original Sampling | Reparameterized Sampling |
|---|---|---|
| Normal | ( z \sim \mathcal{N}(\mu, \sigma^2) ) | ( z = \mu + \sigma \odot \epsilon, \quad \epsilon \sim \mathcal{N}(0, 1) ) [49] |
| Exponential | ( z \sim \text{Exp}(\lambda) ) | ( z = -\frac{1}{\lambda} \log(\epsilon), \quad \epsilon \sim \text{Uniform}(0, 1) ) [49] |
- Implementation: Ensure that the noise variable
ϵis sourced from a fixed distribution and is independent of the model parameters. This allows gradients to be computed with respect toμandσvia backpropagation [49].
The diagram below contrasts the problematic and reparameterized paths for gradient flow:
The Scientist's Toolkit
Research Reagent Solutions for Inflammation Modeling
This table lists key computational "reagents" and methods used in the development and analysis of mathematical models of inflammation, as identified in recent research [8].
| Item | Function in the Context of Inflammation Modeling |
|---|---|
| Lipopolysaccharide (LPS) Data | Used as a calibrated inflammatory stimulus (endotoxin) in both in vitro and human in vivo experiments to elicit a controlled immune response for model development and validation [8]. |
| Ordinary Differential Equation (ODE) Models | The core mathematical framework for describing the dynamics of inflammatory mediators (e.g., cytokines TNF, IL-6, IL-10), immune cell activation, and physiological changes (e.g., heart rate, temperature) over time [8]. |
| Sensitivity Analysis | A computational method to identify which model parameters (e.g., cytokine production rates, mRNA half-lives) have the greatest influence on model outputs, helping to prioritize parameters for estimation or reduction [8]. |
| Profile Likelihood Analysis (PLA) | A method for assessing parameter identifiability by examining how the model's likelihood function changes when a parameter is varied away from its optimal value, confirming that parameters can be uniquely estimated [8]. |
| Fisher Information Matrix Method (FIMM) | A tool for practical identifiability analysis that evaluates whether available data is sufficiently rich to provide precise parameter estimates, helping to diagnose and resolve estimation issues [4]. |
Comparison of Identifiability Analysis Methods
When choosing a method to diagnose parameter identifiability, researchers can refer to the following comparison of common techniques [4].
| Method | Global / Local | Indicator Type | Key Characteristic |
|---|---|---|---|
| DAISY | Both | Categorical | Provides a definitive, theoretical answer for structural identifiability but does not consider real-world data limitations [4]. |
| Sensitivity Matrix Method (SMM) | Local | Both (Categorical & Continuous) | A practical method that analyzes how model outputs change with parameters at specific timepoints in a given dataset [4]. |
| Fisher Information Matrix Method (FIMM) | Local | Both (Categorical & Continuous) | Highly recommended for practical identifiability; can handle random-effects parameters and provides clear, continuous indicators of identifiability strength [4]. |
| Aliasing | Local | Continuous | Scores the similarity between parameters, helping to pinpoint which specific parameters are confounded and causing unidentifiability [4]. |
Enhancing Data Quality and Experimental Design for Better Observability
Frequently Asked Questions (FAQs)
General Graphviz Troubleshooting
Q1: How can I make my Graphviz layout larger?
To increase the size of a layout, you can adjust several individual parameters. Ensure you are not fighting a conflicting graph size setting, like size="6,6", which will scale everything back down [51].
For fdp or neato layouts, increasing the len attribute will expand the layout [51].
Q2: How can I create edges between clusters?
This requires Graphviz version 1.7 or higher. First, set the graph attribute compound=true. Then, you can specify a cluster by name as a logical head or tail to an edge [51].
Q3: How do I generate high-quality, antialiased output? The easiest method is to use vector-based output formats like PDF, SVG, or PostScript. If your Graphviz has a cairo/pango backend, this will also generate antialiased output [51].
Node and Edge Formatting
Q4: How can I change font attributes within a node's label? You can use HTML-like labels to apply different fonts, colors, and sizes within a single node label [52].
Q5: Why are my nodes not filling with color even when style=filled is set?
Ensure you have specified both the style=filled attribute and a fillcolor (or color for some contexts). The command line tools may not always interpret attributes correctly if the graph is heavily pre-processed [53].
Q6: How can I use color schemes for nodes?
The colorscheme attribute allows you to define a namespace for color names. You can then use indices to reference specific colors within that scheme [54].
Graphviz Visualization Guides
The following diagrams provide visual workflows and relationships relevant to experimental design and data observability. All diagrams were created using DOT language and comply with the specified color and contrast guidelines.
Inflammatory Signaling Pathway
Experimental Data Quality Workflow
Identifiability Analysis Process
Research Reagent Solutions
The following table details key materials used in lipid droplet imaging experiments, which are relevant for cellular studies in inflammation research [55].
| Reagent Name | Function/Application | Specific Example |
|---|---|---|
| LD-CPDs (Carbonized Polymer Dots) | Lipid droplet-specific imaging and polarity monitoring [55] | Synthesized from 1,6-dihydroxy naphthalene and ethylenediamine [55] |
| Nile Red | Fluorescent stain for validating lipid droplet targeting [55] | Commercial dye used for correlation analysis [55] |
| 1,6-dihydroxy naphthalene | Hydrophobic precursor for nanoprobe synthesis [55] | Provides benzene ring for lipophilicity [55] |
| Ethylenediamine | Hydrophilic precursor for nanoprobe synthesis [55] | Provides amino group for hydrophilicity [55] |
| RPMI-1640 Medium | Cell culture maintenance for in vitro experiments [55] | Used for growing living cells for imaging [55] |
Table 1: Solvent Polarity vs. Fluorescence Properties of LD-CPDs [55]
| Solvent | Polarity Index | Fluorescence Intensity (a.u.) | Emission Wavelength (nm) |
|---|---|---|---|
| Water | 9.0 | Very Low | Not Reported |
| Acetone | 5.1 | 450 | 540 |
| Dichloromethane | 3.1 | 850 | 520 |
| n-Hexane | 0.0 | 1200 | 505 |
Table 2: Key Experimental Parameters for Lipid Droplet Imaging
| Parameter | Optimal Value/Method | Purpose/Outcome |
|---|---|---|
| Synthesis Temperature | 160°C | Optimal fluorescence intensity of LD-CPDs [55] |
| Pearson's Correlation Coefficient | 0.95 with Nile Red | Validates specificity for lipid droplets [55] |
| Incubation Time | 4 hours | Sufficient for cellular uptake and wash-free imaging [55] |
| Viability Assay | >90% | Confirms low cytotoxicity of LD-CPDs [55] |
Addressing Local Solutions and Bimodality in Parameter Estimation
This technical support center provides troubleshooting guides and FAQs for researchers, scientists, and drug development professionals working with mathematical models in inflammation research. The content is framed within a broader thesis on resolving identifiability issues to ensure reliable parameter estimation and model predictions.
# Core Concepts: Identifiability in Inflammation Modeling
FAQ: What is the difference between structural and practical identifiability, and why does it matter for my model of neuroinflammation?
- Structural Identifiability is a theoretical property of your model. It investigates whether model parameters can be uniquely determined from ideal, noise-free infinite amount of data. A model that is not structurally identifiable cannot yield unique parameter estimates even under perfect conditions due to its mathematical structure [14] [56]. This is often a result of parameter correlations [18].
- Practical Identifiability investigates the ability of a model to reveal its parameters under real-world conditions: scarce and noisy data. A model can be structurally identifiable but not practically identifiable if the available data is insufficient to pinpoint parameter values accurately [14] [20].
Neglecting these analyses can lead to unreliable parameter estimates, resulting in ambiguous or misleading biological conclusions and potentially misguided intervention strategies [18] [56].
FAQ: What are "local solutions" and "bimodality" in the context of parameter estimation?
- Local Solutions refer to parameter sets that provide a good fit to the data within a limited region of the parameter space but may not represent the best possible fit (global optimum). Optimization algorithms can get "stuck" in these local solutions [56].
- Bimodality in parameter estimation occurs when the objective function (e.g., the measure of fit between the model and data) has two distinct, well-separated parameter sets that fit the data equally well. This is a strong indicator of non-identifiability and suggests that the available data cannot distinguish between two different biological mechanisms or parameter interpretations [56].
# Troubleshooting Guides
Guide 1: Diagnosing and Resolving Local Solution Convergence
Problem: Your optimization algorithm consistently converges to different parameter values depending on the initial guess, suggesting trapping in local solutions.
Diagnostic Steps:
- Check Structural Identifiability: Before fitting, verify your model is structurally identifiable using tools like
StructuralIdentifiability.jlin Julia or DAISY [18]. This rules out fundamental mathematical issues. - Profile Likelihood Analysis: Perform a profile likelihood analysis for each parameter. A flat profile indicates practical non-identifiability, while a profile with multiple peaks suggests local solutions or bimodality [56].
- Multi-Start Optimization: Run your estimation routine from a wide variety of initial parameter guesses. Clustering of results into several distinct groups indicates local solutions.
Solutions:
- Refine Experimental Design: Ensure data is collected during dynamic phases of the system response, not just at steady state. Data from early time points is often critical [14] [20].
- Incorporate Additional Data Types: Fit your model to multiple types of data simultaneously. For a neuroinflammation model, this could include time-course data for different species like cytokines, immune cell counts, and neuronal health markers [14] [57].
- Use Global Optimization Algorithms: Employ metaheuristics like evolutionary algorithms or particle swarm optimization, which are better at escaping local minima than local search methods [56].
The following workflow outlines the diagnostic process:
Guide 2: Managing Bimodality and Practical Non-Identifiability
Problem: Your analysis reveals two distinct sets of parameters that fit your data equally well (bimodality), or parameter confidence intervals are extremely wide (practical non-identifiability).
Diagnostic Steps:
- Correlation Analysis: Calculate the correlation matrix of parameter estimates. Parameters with correlations near +1 or -1 are often non-identifiable and can form a bimodal distribution in the parameter space [14] [56].
- Posterior Distribution Inspection: If using Bayesian methods, inspect the marginal posterior distributions of parameters. Bimodal distributions will be directly visible.
- Model Selection: Test if a simplified model, with fewer parameters or a different structure, can describe the data without exhibiting bimodality.
Solutions:
- Parameter Fixing: If certain parameters are known from prior literature, fix them to a constant value to reduce the dimensionality of the estimation problem [14].
- Model Reparameterization: Combine non-identifiable parameters into a single identifiable composite parameter. For example, in a simple viral infection model
dI/dt = βTV - δI, if only viral loadVis observed, the productπβmight be identifiable whereas the individual parametersπandβare not [14] [18]. - Optimal Experimental Design: Design new experiments to collect data that specifically targets the non-identifiable parameters. This often involves perturbing the system or measuring at optimal time points to maximize information gain [20].
# Advanced Protocols
Protocol: A Workflow for Robust Parameter Estimation in Complex Inflammation Models
This protocol provides a detailed methodology for ensuring reliable parameter estimation, integrating concepts from the cited literature [14] [18] [56].
I. Pre-Fitting Analysis
- Structural Identifiability Check: Using your model equations and defined observables (e.g., cytokine concentrations), run an analysis with
StructuralIdentifiability.jl[18]. - Sensitivity Analysis: Perform a global sensitivity analysis (e.g., Sobol indices) to determine which parameters most influence model outputs. Focus estimation efforts on these sensitive parameters [56].
II. Data Preparation and Integration
- Leverage All Data: Collate all available quantitative data, including time-course measurements for multiple model variables if possible (e.g., viral titer and immune cell data) [14].
- Weight Data Appropriately: Account for differences in scale and uncertainty between different types of measurements (e.g., RNA copies vs. cell counts).
III. Estimation and Validation
- Global Optimization with Multi-Start: Use a global optimizer from a wide range of initial parameter values to thoroughly explore the parameter space [56].
- Practical Identifiability Assessment: Calculate profile likelihoods or Markov Chain Monte Carlo (MCMC) samplings to establish reliable confidence intervals for the estimated parameters [14] [20].
- Cross-Validate: Where data allows, use a subset of data for model calibration and the remaining data for validation.
The workflow for this protocol is visualized below:
# Research Reagent Solutions
The following table details key computational tools and their functions for addressing identifiability and estimation problems.
| Research Tool / Reagent | Function / Explanation |
|---|---|
| StructuralIdentifiability.jl [18] | A Julia package for assessing structural identifiability of ODE models using a differential algebra approach. It can handle complex, high-dimensional models. |
| DAISY Software [18] [20] | A differential algebra tool used for structural identifiability analysis, often used for validation and comparison. |
| Profile Likelihood Analysis [56] | A methodology for assessing practical identifiability by examining how the model's fit changes as a parameter is fixed away from its optimal value. |
| Global Optimizers [56] | Algorithms (e.g., evolutionary, particle swarm) designed to search the entire parameter space to find the global optimum and avoid local solutions. |
| Multi-Start Optimization [56] | A strategy involving running a local optimizer from many starting points to map the topography of the objective function and identify local/global solutions. |
| Two-Patch Within-Host Model [20] | A model structure that incorporates spatial or physiological heterogeneity (e.g., upper/lower respiratory tract) to improve parameter identifiability by providing more information. |
Frequently Asked Questions
Q1: Why should I replace bilinear terms in my within-host model? Bilinear (or mass-action) terms, often of the form ( \beta V C ) for virus-cell interactions, are a common source of structural non-identifiability. These terms assume that interaction rates can increase indefinitely. However, real biological processes, such as the cytotoxic T lymphocyte (CTL)-driven elimination of infected cells or virus-induced CTL expansion, are saturable. Refining these to bounded-rate functions (e.g., Michaelis-Menten) provides a more biologically realistic description and can resolve non-identifiability by decoupling parameter influences, leading to more reliable and interpretable parameter estimates [58].
Q2: What is the difference between structural and practical identifiability?
- Structural Identifiability is a theoretical property of your model. A parameter is structurally identifiable if it can be uniquely determined from perfect, noise-free experimental data. It depends only on the model's equations and what is being observed [59].
- Practical Identifiability is a more pragmatic concern. A parameter is practically identifiable if it can be estimated with sufficient confidence from the limited, noisy data that is actually available. A model can be structurally identifiable but not practically identifiable if the data is insufficient in quality or quantity [59].
Q3: My model fits the data well but parameters have wide confidence intervals. What does this mean? This is a classic sign of practical non-identifiability. Your model can reproduce the observed data for a wide range of different parameter combinations. This often occurs when parameters are correlated, meaning a change in one can be compensated for by a change in another without affecting the goodness-of-fit. This indicates that your data does not contain enough information to uniquely pin down all parameter values, and predictions outside the fitted conditions may be unreliable [59].
Q4: How can I assess the identifiability of my model? You can use a combination of differential algebraic techniques for structural identifiability and a Bayesian approach or a profile likelihood analysis for practical identifiability. In a profile likelihood analysis, you fix a parameter to a series of values and re-optimize all other parameters. A flat profile indicates a non-identifiable parameter, while a well-defined minimum suggests identifiability [58] [59].
Q5: How does model complexity relate to identifiability? Increasing model complexity by adding more mechanisms or parameters generally improves the model's ability to fit data (goodness-of-fit). However, it often makes parameters less identifiable because a change in one parameter can be more easily compensated for by changes in other parameters. Therefore, identifiability should be considered alongside goodness-of-fit and complexity during model selection [59].
Troubleshooting Guides
Problem: Poor Convergence During Parameter Estimation
Symptoms: Parameter estimation algorithms fail to converge, or results are highly sensitive to initial guesses.
Potential Causes and Solutions:
| Cause | Solution |
|---|---|
| Structurally non-identifiable parameters | Perform a structural identifiability analysis using software like StructuralIdentifiability.jl. Reparameterize or simplify the model to eliminate non-identifiable parameters [58]. |
| Overly complex model for available data | Simplify the model by reducing the number of parameters or fixing well-established values from literature. Use model selection criteria (e.g., AIC, BIC) that balance complexity with goodness-of-fit [59]. |
| Poor-quality or insufficient data | The model may require data with higher resolution or from multiple observable outputs (e.g., viral load and immune cell counts). Design experiments to provide dynamic data that captures key transitions [60]. |
Problem: Biologically Implausible Parameter Estimates
Symptoms: Fitted parameters have values that are orders of magnitude outside expected physiological ranges.
Potential Causes and Solutions:
| Cause | Solution |
|---|---|
| Incorrect model structure | The model may lack a key biological mechanism. Review the underlying biology; for instance, replace a bilinear incidence term ( \beta V T ) with a bounded, saturable function like a Michaelis-Menten term ( \frac{\beta V T}{K + V} ) [58]. |
| Compensation between parameters | Perform a sensitivity or identifiability analysis to detect correlated parameters. Consider fixing one of the correlated parameters to a literature value or re-parameterizing the model to combine them into a single, identifiable lumped parameter [59]. |
Problem: Model Fails to Predict New Experimental Outcomes
Symptoms: The model calibrated to one dataset fails to accurately predict the system's behavior under different conditions or treatments.
Potential Causes and Solutions:
| Cause | Solution |
|---|---|
| Practical non-identifiability | A model with non-identifiable parameters may fit one dataset but lacks predictive power. Conduct a practical identifiability analysis using profile likelihoods to ensure all parameters are well-constrained by the data [59]. |
| Lack of key biological dynamics | The model might be missing a critical feedback loop or regulatory mechanism. For example, in immune response modeling, ensure important components like macrophage polarization (M1/M2) or the anti-inflammatory response (e.g., IL-10) are included if relevant [61]. |
Experimental Protocols for Identifiability Analysis
Protocol 1: Profile Likelihood for Practical Identifiability
This protocol assesses how well a parameter can be identified from a specific dataset [59].
- Calibrate the Model: Find the maximum likelihood estimate (MLE) for the full parameter vector ( \hat{\theta} ).
- Profile a Parameter: Select a parameter of interest, ( \thetai ). Define a range of values for ( \thetai ) around its MLE.
- Re-optimize: For each fixed value of ( \theta_i ) in the range, re-optimize the likelihood function over all other free parameters.
- Calculate Profile Likelihood: For each value of ( \theta_i ), calculate the optimized profile likelihood value.
- Analyze Profile: A parameter is practically identifiable if its profile likelihood plot shows a well-defined, unique minimum. A flat profile indicates the parameter is not identifiable from the data.
Protocol 2: Model Selection Based on Identifiability
This protocol helps select the most appropriate model from a set of candidates [59].
- Define Candidate Models: Develop a set of models of varying complexity that describe the same biological phenomenon (e.g., Fisher-KPP model and its extensions for cell invasion).
- Calibrate and Assess Fit: Calibrate each model to the experimental data and record the goodness-of-fit (e.g., residual sum of squares).
- Perform Identifiability Analysis: Conduct a practical identifiability analysis (as in Protocol 1) for each model.
- Select Model: The preferred model should have a good balance of an acceptable goodness-of-fit and a high proportion of identifiable parameters. A model with a slightly worse fit but all identifiable parameters is often preferable to a better-fitting model with non-identifiable parameters.
The table below summarizes key properties of different incidence rate functions used in within-host models, highlighting their impact on identifiability.
| Incidence Rate Function | Mathematical Formulation | Key Characteristics | Impact on Identifiability |
|---|---|---|---|
| Bilinear (Mass-Action) | ( \beta V T ) | Linear, unbounded growth with pathogen and target cell density. | Often leads to structural non-identifiability as parameters like the infection rate ( \beta ) and production rate ( p ) can be correlated [58]. |
| Michaelis-Menten (Holling Type II) | ( \frac{\beta V T}{K + V} ) | Bounded, saturable rate. Accounts for limited resources or processing time. | Improves identifiability by introducing a half-saturation constant ( K ), which helps decouple parameter influences [58] [60]. |
| Beddington-DeAngelis | ( \frac{\beta V T}{1 + \alpha1 T + \alpha2 V} ) | Bounded rate that accounts for interference by both target cells and free virus. | Can improve identifiability by more accurately capturing complex interactions, but may require more data to identify the additional parameters ( \alpha1 ) and ( \alpha2 ) [60]. |
Model Transition and Experimental Workflow
The following diagram illustrates the logical process of refining a model from a bilinear to a bounded-rate structure and the subsequent steps for validation.
Research Reagent Solutions
The table below lists key computational tools and their functions for addressing identifiability in mathematical immunology.
| Research Tool | Function in Identifiability Analysis | Key Features / Use Case |
|---|---|---|
| StructuralIdentifiability.jl [58] | A Julia-based package for assessing structural identifiability of ODE models. | Uses differential algebraic techniques to determine if parameters can be uniquely identified from perfect data. |
| Profile Likelihood Analysis [59] | A computational method for assessing practical identifiability. | Evaluates parameter identifiability from real, noisy data by analyzing the shape of the likelihood profile. |
| DynamicHMC.jl [58] | A Julia-based package for Bayesian parameter inference using Hamiltonian Monte Carlo (HMC). | Useful for parameter estimation and exploring parameter uncertainties in complex, high-dimensional models. |
| AIC / BIC [59] | Information criteria used for model selection. | Helps balance model goodness-of-fit against complexity; a simpler model with identifiable parameters is often preferred. |
A Posteriori Identifiability Analysis and Confidence Interval Estimation
Frequently Asked Questions (FAQs)
Q1: What is the difference between structural and practical (a posteriori) identifiability?
- Structural Identifiability is a theoretical property of your model. It asks whether model parameters can be uniquely identified assuming perfect, noise-free experimental data. It is a prerequisite for practical identifiability analysis [21] [14].
- Practical (A Posteriori) Identifiability assesses whether parameters can be reliably estimated given the limitations of your real-world data, which is often noisy, sparse, and limited in scope. It is performed after parameter estimation to evaluate the reliability of your estimates [21] [44] [14].
Q2: My model parameters are not practically identifiable. What are the main causes?
Non-identifiability typically arises from three main areas [14]:
- Model Structure: Over-parameterization or high correlations between parameters.
- Data Issues: Data that is too noisy, insufficient in quantity, or lacks information on key model variables.
- Experimental Design: Measurement times that do not capture critical dynamics of the system.
Q3: What are the common methods for assessing practical identifiability?
Two widely used methods are:
- Profile Likelihood Analysis: This method involves varying one parameter while re-optimizing all others to see how the model's fit changes. A flat profile indicates a non-identifiable parameter [8] [21].
- Fisher Information Matrix (FIM) Analysis: The FIM is calculated from the model output sensitivities. If the FIM is invertible, parameters are considered practically identifiable. Eigenvalue decomposition of the FIM can reveal which parameter combinations are identifiable [21].
Troubleshooting Guides
Guide 1: Resolving Non-Identifiable Parameters
Follow this systematic procedure to diagnose and address identifiability issues [62] [63]:
Step 1: Identify the Problem
- Confirm that parameter estimates have unacceptably large confidence intervals or that different initial guesses lead to very different parameter values.
Step 2: List Possible Causes
- Data-Related: Insufficient data points, lack of data for key model variables, poor data quality (high noise), or non-informative measurement timepoints.
- Model-Related: Model is too complex (over-parameterized), existence of practically redundant parameters, or incorrect model structure.
Step 3: Collect Data & Diagnose
Step 4: Eliminate Causes & Experiment
- If data is the issue: Consider if you can collect more data, especially for poorly observed variables, or at timepoints that capture system dynamics better. Optimal experimental design algorithms can help here [21].
- If the model is the issue: Consider simplifying the model by fixing non-identifiable parameters to literature values, or by re-parameterizing the model to reduce correlation.
Step 5: Implement Solution & Verify
- Implement the chosen fix (e.g., collect new data, simplify model).
- Re-run the identifiability analysis to confirm that parameters are now identifiable with tighter confidence intervals.
Guide 2: Workflow for Reliable Parameter Estimation & CI Calculation
This workflow integrates model calibration, identifiability checking, and confidence interval estimation for robust results [44].
Key Methodologies and Protocols
Profile Likelihood Analysis
This is a powerful method for testing the practical identifiability of individual parameters.
Detailed Protocol:
- Estimate Parameters: Find the parameter vector ( \hat{\theta} ) that provides the best fit to your experimental data, minimizing a cost function ( C(\theta) ) (e.g., sum of squared errors).
- Profile a Parameter: Select a parameter of interest, ( \thetai ). For a series of fixed values ( \thetai = g ) around its optimal estimate ( \hat{\theta}i ), re-optimize the cost function ( C(\theta) ) over all other parameters ( \theta{j \neq i} ).
- Calculate Profile Likelihood: For each value ( g ), record the optimized value of the cost function. The profile likelihood for ( \thetai ) is defined as ( PL(\thetai) = \min{\theta{j \neq i}} C(\theta) ).
- Evaluate the Profile: Plot ( PL(\theta_i) ) against the values of ( g ). A uniquely identifiable parameter will show a clear, V-shaped minimum. A flat or shallow profile indicates that the parameter is not practically identifiable from the data [8] [21].
Confidence Interval Estimation via Fisher Information Matrix (FIM)
For parameters that are deemed identifiable, the FIM provides a way to estimate confidence intervals.
Detailed Protocol:
- Compute the FIM: The FIM, ( F(\hat{\theta}) ), is calculated based on the model's output sensitivities at the optimal parameter estimate ( \hat{\theta} ). The sensitivity matrix ( s(\theta) ) is defined by the derivatives of model outputs with respect to parameters. The FIM is given by ( F(\hat{\theta}) = s^T(\hat{\theta}) s(\hat{\theta}) ) [21].
- Invert the FIM: The covariance matrix ( C ) of the parameter estimates is approximated by the inverse of the FIM: ( C \approx F^{-1}(\hat{\theta}) ).
- Calculate Confidence Intervals: The standard error for parameter ( \thetai ) is the square root of the corresponding diagonal element of the covariance matrix: ( \text{se}(\thetai) = \sqrt{C{ii}} ). The approximate ( 95\% ) confidence interval is then ( \hat{\theta}i \pm 1.96 \cdot \text{se}(\theta_i) ) [21] [44].
The Scientist's Toolkit: Research Reagent Solutions
Table 1: Essential Computational Tools for Identifiability Analysis
| Tool/Reagent | Function/Benefit |
|---|---|
| Profile Likelihood | Diagnoses identifiability of individual parameters; reveals flat relationships that indicate non-identifiability [8] [21]. |
| Fisher Information Matrix (FIM) | Assesses overall parameter identifiability; its invertibility is key. Eigenvalue decomposition identifies identifiable parameter combinations [21]. |
| Optimal Experimental Design Algorithms | Determines the most informative timepoints for data collection to ensure parameter identifiability, maximizing the utility of experiments [21]. |
| Hybrid Neural ODEs (HNODEs) | Combines mechanistic models with neural networks to represent unknown system components, allowing parameter estimation even with incomplete mechanistic knowledge [44]. |
| Regularization Techniques | Adds constraints to the model fitting process to handle non-identifiable parameters and improve numerical stability during estimation [21]. |
Workflow for Inflammation Research Context
The following diagram outlines a specific computational workflow for applying these concepts to mathematical models of inflammation, such as those involving LPS exposure [8] [44].
Benchmarking and Validation: Ensuring Model Reliability for Clinical Translation
In mathematical models of inflammation, structural identifiability determines if unique parameter values can be found from ideal noise-free data, while practical identifiability assesses whether this is feasible with real, noisy experimental data. Parameters like K1, which often represent synthesis rates, degradation constants, or activation thresholds, frequently suffer from non-identifiability, where different parameter combinations yield identical model outputs. This directly impedes our ability to correlate computational predictions with biologically meaningful endpoints, as an unidentifiable parameter cannot be reliably used to draw conclusions about biological mechanisms. Resolving these issues is therefore not merely a mathematical exercise but a critical step in ensuring model predictions have translational value for drug development.
Troubleshooting Guides
Guide: Diagnosing and Resolving Parameter Non-Identifiability
Problem: Model parameters (e.g., K1) cannot be uniquely determined from experimental data, leading to unreliable correlations with biological endpoints.
Symptoms:
- Large confidence intervals on parameter estimates from fitting procedures.
- Strong correlations between different parameter values (e.g., between a production rate and a degradation rate).
- Small changes in data leading to large, unpredictable shifts in estimated parameter values.
- Failure of the model to generalize to new experimental conditions.
Solution Steps:
Profile Likelihood Analysis: This is the gold standard for assessing practical identifiability.
- Procedure: a. For a parameter of interest (e.g., K1), fix its value across a defined range. b. At each fixed value, optimize the log-likelihood function by allowing all other model parameters to vary. c. Plot the optimized log-likelihood against the parameter value. A uniquely identifiable parameter will show a clearly defined minimum (a V-shaped profile). A flat or shallow profile indicates non-identifiability [8].
- Example: A study modeling the inflammatory response to lipopolysaccharide (LPS) used profile likelihood analysis to confirm the unique identifiability of six sensitive parameters, including mRNA half-lives and scaling factors, which were subsequently estimated using calibration data [8].
Sensitivity Analysis: Determine which parameters most significantly influence model outputs that correspond to measurable biological endpoints.
- Procedure: Calculate local (e.g., using partial derivatives) or global (e.g., using Sobol' indices or Morris method) sensitivity measures. Parameters with low sensitivity are often practically non-identifiable and may be candidates for fixing to a literature value [8] [9].
- Example: A mathematical model of osteoarthritis inflammation used local sensitivity analysis and bifurcation diagrams to verify the model and evaluate the regulatory mechanism of inflammation dynamics by adipokines [9].
Model Reparameterization: Reduce parameter interdependence.
- Action: If two parameters, K1 and K2, are highly correlated, consider combining them into a single composite parameter (e.g., K1/K2) that may be more identifiable.
Incorporating Additional Data Types: Constrain parameters by fitting the model to diverse datasets.
- Action: If K1 is non-identifiable from cytokine time-series data alone, incorporate data from a different biological endpoint, such as immune cell counts or tissue damage markers [64] [65]. A model of innate immunity to inhaled toxicants was developed and calibrated in stages, first using in vitro data from human lung epithelial cells and then data from mouse-derived macrophages, which helped constrain different parameter sets [65].
Guide: Validating Model Correlations with Biological Endpoints
Problem: A model parameter (K1) shows a statistically significant correlation with an experimental endpoint (e.g., inflammatory cell influx), but the relationship lacks biological plausibility.
Symptoms:
- The correlation is strong in the training data but vanishes in validation data.
- The direction of the correlation contradicts established biological knowledge (e.g., a putative anti-inflammatory mediator correlates with increased inflammation).
- The correlation is driven by outliers in the dataset.
Solution Steps:
Multi-Scale Model Validation: Ensure the model is validated against multiple, orthogonal endpoints.
- Procedure: A model predicting that K1 correlates with macrophage activation should be tested not just against cytokine levels (the same level of biology), but also against histology scores for tissue inflammation or clinical pain scores [66]. For instance, a 3D potency assay for a cell therapy in osteoarthritis correlated in vitro secretory profiles with the clinical endpoints of VAS and KOOS pain scores after 12 months, providing a robust, multi-scale validation [66].
Cross-Validation: Assess the robustness of the correlation.
- Procedure: Use k-fold or leave-one-out cross-validation to determine if the K1-endpoint relationship holds across different subsets of the data. This helps identify spurious correlations caused by overfitting [66].
Causal Inference Analysis: Investigate whether the relationship between K1 and the endpoint is likely to be causal.
- Procedure: Techniques like Mendelian randomization can be used with human genetic data to support causal relationships. One study integrated Mendelian randomization with multi-omics to identify TNIK as a key causal gene in gut microbiota-induced IBD development, moving beyond mere correlation [67].
Frequently Asked Questions (FAQs)
Q1: My model fits the training data well, but the estimated value for K1 varies wildly between experimental replicates. What is the most likely cause? A: This is a classic sign of practical non-identifiability. The profile likelihood for K1 is likely to be flat or very shallow, meaning the data do not contain sufficient information to pin down its value uniquely. Follow the troubleshooting guide above, focusing on profile likelihood analysis and incorporating additional data types to better constrain K1.
Q2: Are there specific types of experimental data that are particularly effective for constraining inflammatory model parameters? A: Yes. Time-series data is vastly more informative than single time-point measurements for dynamic models [8] [68]. Data that captures the peak and resolution phases of inflammation are crucial for distinguishing between pro-inflammatory and anti-inflammatory parameters. Furthermore, measuring multiple interconnected variables (e.g., cytokines, immune cell populations, and tissue damage markers) provides cross-constraints that greatly improve identifiability [64] [65] [9]. For example, simultaneously measuring TNF-α, IL-6, and IL-10 can help separate production and inhibition parameters.
Q3: How can I handle a parameter like K1 that is sensitive (so it's important) but non-identifiable (so its value is uncertain)? A: This is a common challenge. Your options are:
- Design a new experiment specifically to target the uncertainty in K1, such as a perturbation experiment (e.g., a knockout or inhibitor study).
- Fix the parameter to a value from the literature obtained from a more reductionist experimental setup (e.g., an in vitro assay).
- Report the uncertainty transparently. Use confidence intervals or Bayesian posteriors to show the plausible range of K1 and propagate this uncertainty to model predictions, making it clear that the correlation with the biological endpoint exists within a range of K1 values.
Q4: In the context of a complex, multi-scale inflammation model, what does a "biological endpoint" refer to? A: A biological endpoint is a measurable indicator of a biological state or process. In inflammation research, these exist at multiple scales:
- Molecular: Cytokine concentrations (e.g., TNF-α, IL-6) [8] [65], complement activation fragments (e.g., C3dg) [69].
- Cellular: Immune cell counts (e.g., neutrophil infiltration, M1/M2 macrophage ratio) [65] [61], microglial activation [70].
- Tissue: Histopathology scores, tissue damage markers, fibrosis [65] [70].
- Clinical/Systemic: Disease severity scores (e.g., Mayo score for IBD [67]), pain scores (e.g., VAS for osteoarthritis [66]), survival.
Experimental Protocols for Key Workflows
Protocol: Profile Likelihood Analysis for Practical Identifiability
Objective: To empirically determine the practical identifiability of a parameter (e.g., K1) in a mathematical model of inflammation.
Materials:
- A calibrated mathematical model (ODE/PDE system).
- Experimental time-course data for model fitting.
- Computing environment (e.g., MATLAB, R, Python).
Method:
- Model Calibration: Perform an initial fit of the model to the data to obtain a nominal parameter set, including K1â‚€.
- Define Parameter Range: Define a realistic range for K1 around K1₀ (e.g., ± two orders of magnitude).
- Profile Calculation: a. Discretize the K1 range into N points. b. For each point K1áµ¢ in the range: i. Fix the parameter K1 at the value K1áµ¢. ii. Optimize the log-likelihood function by varying all other free parameters in the model. iii. Record the optimized log-likelihood value.
- Visualization and Interpretation: Plot the optimized log-likelihood values against the K1 values. A uniquely identifiable parameter will exhibit a quadratic-like, V-shaped profile with a clear minimum. A flat profile indicates non-identifiability [8].
Protocol: IntegratingIn VitroandIn SilicoData for Model Calibration
Objective: To constrain model parameters by leveraging data from reductionist in vitro experiments before fitting to complex in vivo data.
Materials:
- In vitro experimental system (e.g., primary immune cells, cell lines).
- Assays for relevant endpoints (ELISA for cytokines, flow cytometry for cell markers).
- Mathematical model.
Method:
- Submodel Development: Decompose the full model into simpler submodels that represent specific processes measurable in vitro. For example, develop an "epithelial-cytokine" submodel [65].
- In Vitro Experiment: Expose the in vitro system to a controlled inflammatory stimulus (e.g., LPS, wood smoke particulate matter [65]). Collect time-series data on key outputs (e.g., pro-inflammatory cytokine secretion).
- Submodel Calibration: Fit the submodel exclusively to the in vitro data to estimate a subset of the parameters. This provides well-justified fixed values for these parameters in the full model.
- Full Model Calibration: Using the parameters fixed from in vitro work, calibrate the remaining parameters of the full model against in vivo or more complex ex vivo data. This sequential calibration reduces the number of simultaneously fitted parameters, mitigating identifiability issues [65].
Signaling Pathways and Workflow Diagrams
Core Inflammation Network
Diagram Title: Core Inflammation Regulatory Network
Identifiability Analysis Workflow
Diagram Title: Parameter Identifiability Assessment Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table: Essential Reagents for Inflammation Model Validation
| Reagent / Material | Function in Experiment | Example Application |
|---|---|---|
| Lipopolysaccharide (LPS) | Pathogen-associated molecular pattern (PAMP) used to induce a standardized, acute inflammatory response in vivo (experimental endotoxemia) and in vitro [8]. | Calibrating models of systemic inflammatory response, such as sepsis [8]. |
| PEG-4MAL Hydrogel | A synthetic, tunable hydrogel used for 3D cell encapsulation in microfluidic "organ-on-a-chip" devices. Provides a more physiologically relevant microenvironment than 2D culture [66]. | Creating 3D potency assays to predict clinical efficacy of cell therapies (e.g., for osteoarthritis) by measuring secretory profiles [66]. |
| Simulated Synovial Fluid (simSF) | A formulated culture medium mimic containing the most abundant proteins and glycosaminoglycans found in osteoarthritic synovial fluid. Used to test cell response in a disease-relevant milieu [66]. | Evaluating the secretory response of bone marrow aspirate concentrate (BMAC) cells in a clinically predictive potency assay [66]. |
| Pattern Recognition Molecule (PRM) Assays | Immunoassays (e.g., TRIFMA, ELISA) to quantify plasma levels of PRMs (e.g., MBL, Ficolins) and complement activation fragments (e.g., C3dg) [69]. | Measuring biomarkers of innate immune activation and dysregulation to correlate with infection risk in chronic diseases like CKD [69]. |
| AAV9 Viral Vectors | Adenovirus-associated virus serotype 9, used for efficient in vivo gene delivery and overexpression in animal models. | Validating candidate genes (e.g., TNIK in IBD) by modulating their expression and observing the effect on disease severity and inflammatory endpoints [67]. |
Mathematical modeling is an indispensable tool for understanding the complex dynamics of the inflammatory response, a process characterized by a sophisticated interplay of immune cells, signaling molecules, and tissue damage [61]. Inflammatory Bowel Disease (IBD), for instance, involves host genetic predisposition, gut microbial dysbiosis, and immunological inconsistencies, making it a prime candidate for computational analysis [71]. Researchers employ various modeling frameworks—Ordinary Differential Equations (ODEs), Partial Differential Equations (PDEs), and Boolean Networks—to capture different aspects of these biological systems. Each paradigm offers distinct advantages and suffers from specific limitations, particularly concerning identifiability issues, where model parameters cannot be uniquely determined from available experimental data. This technical support guide provides a comparative analysis of these approaches, complete with troubleshooting advice and experimental protocols, to aid researchers in selecting and implementing the most appropriate model for their specific research questions in inflammation and beyond.
Core Definitions and Applications
- Ordinary Differential Equations (ODEs) are used to model the time-dependent evolution of system species concentrations (e.g., cytokines, immune cells) within a well-mixed environment. They are defined by equations of the form ( y′(t)=f(y,p) ), where the rate of change of a species depends on the current state of the system and a set of parameters [72]. ODEs are a cornerstone of classical systems biology for modeling metabolic and signaling pathways [73].
- Partial Differential Equations (PDEs) extend this framework to include spatial heterogeneity, modeling how concentrations change over both time and space. They are crucial for capturing processes like immune cell infiltration into tissue or the formation of granulomas in infections [61]. A general form is ( ∂_t u(t,x) + ℒu = f(t,x) ), where ( ℒ ) is a spatial differential operator [74].
- Boolean Networks represent a qualitative, discrete modeling approach. Each variable (e.g., a gene or protein) is represented as a node that can be in one of two states: ON (1) or OFF (0). The state of a node at the next time step is determined by a logical rule (Boolean function) based on the states of its inputs [75]. This formalism is powerful for modeling large-scale regulatory and signaling networks where detailed kinetic parameters are unavailable [76].
Comparative Analysis: Strengths, Weaknesses, and Ideal Use Cases
Table 1: High-level comparison of ODE, PDE, and Boolean Network modeling paradigms.
| Feature | Ordinary Differential Equations (ODEs) | Partial Differential Equations (PDEs) | Boolean Networks |
|---|---|---|---|
| System Representation | Continuous, quantitative, time-dependent | Continuous, quantitative, time- and space-dependent | Discrete, qualitative, state-based |
| Biological Interpretation | Captures quantitative dynamics and rates | Captures spatiotemporal dynamics and gradients | Captures logical structure and necessity/sufficiency of interactions |
| Typical Applications | Signaling pathways, metabolic kinetics, population dynamics | Tissue-scale inflammation, wound healing, morphogenesis | Gene regulatory networks, signaling logic, large-scale network analysis [76] |
| Data Requirements | Quantitative time-series data for parameter estimation | Quantitative time-series and spatial data | Network topology, qualitative knowledge of activating/inhibiting interactions |
| Handling of Identifiability | Prone to identifiability issues with many unknown parameters [73] | Highly prone to identifiability issues due to increased complexity | Avoids kinetic parameter identifiability by abstracting to logic |
| Computational Complexity | Moderate to high (depends on stiffness and size) | High to very high | Low, enables analysis of genome-scale networks [73] |
| Key Advantage | Quantitative precision and dynamic prediction | Resolution of spatial heterogeneity | Scalability and intuitive logic in the face of uncertainty |
Section 2: Technical Deep Dive and Troubleshooting
Frequently Asked Questions (FAQs)
Q1: My ODE model of the NF-κB pathway has many unknown parameters, leading to poor identifiability. What are my options? A1: You have several paths forward:
- Incorporate Prior Knowledge: Use literature to fix certain parameters or constrain their plausible ranges.
- Model Reduction: Simplify the model by quasi-steady-state assumptions or by lumping parameters, reducing the total number to be estimated.
- Switch Paradigms: If quantitative predictions are not essential, consider translating your ODE model into a Boolean Network to focus on the essential logic and topology of the pathway, which inherently avoids kinetic parameter identifiability issues [72].
- Scientific Machine Learning (SciML): Explore hybrid approaches that combine mechanistic ODE models with machine learning. ML can be used to learn parts of the model where parameters are unknown, constrained by the known physics of the ODE framework [73].
Q2: When should I choose a Boolean model over a more precise ODE model? A2: A Boolean model is the superior choice when:
- The available biological data is qualitative (e.g., "gene X is expressed" or "protein Y is active") rather than quantitative.
- The system you are modeling is very large (e.g., a whole-genome regulatory network), making the specification and parameterization of ODEs infeasible [73].
- The primary research question revolves around the logical structure of the network, such as identifying key regulators, essential pathways, or the stable attractor states (e.g., cell fates) of the system [75] [76].
Q3: How can I validate a Boolean model if it doesn't produce quantitative outputs? A3: Boolean models are validated against qualitative experimental outcomes.
- Stable States: Check if the model's point attractors (steady states) correspond to known biological phenotypes (e.g., proliferation, apoptosis, differentiation) [72].
- Perturbation Analysis: Simulate knock-outs (node fixed to 0) or over-expressions (node fixed to 1) and verify if the model's response matches experimental observations (e.g., expected changes in cell fate or marker expression) [75].
- Sequence of Activation: Ensure that the temporal sequence of node activation in asynchronous Boolean simulations aligns with known pathway activation patterns.
Q4: My PDE model is computationally prohibitive to simulate. How can I make it more tractable? A4:
- Dimensionality Reduction: Exploit model symmetries to reduce the spatial dimensions (e.g., from 3D to 2D or 1D) where possible.
- Advanced Discretization: Use adaptive mesh refinement to concentrate computational effort only in regions with high activity gradients.
- Operator Learning: Consider using modern surrogate models like DeepONet or NODE-ONet [77] [74]. These neural network-based models learn the mapping from input functions (e.g., initial conditions, parameters) to output solutions. Once trained, they can predict solutions almost instantaneously, bypassing the need for expensive numerical solvers for new simulations.
Troubleshooting Common Modeling Issues
Table 2: Common issues, their likely causes, and potential solutions across modeling paradigms.
| Problem | Likely Cause | Potential Solution |
|---|---|---|
| ODE instability/divergence | Model stiffness; Poorly chosen numerical solver; Incorrect parameter sets. | Use a stiff solver (e.g., Rodas5P(), CVODE_BDF [78]); reduce step size; check parameter units and magnitudes. |
| Poor ODE fit to data | Structural non-identifiability; Over-parameterization; Incorrect model structure. | Perform identifiability analysis; fix or remove unidentifiable parameters; simplify model; consider a different biological hypothesis. |
| Boolean model gets stuck in unrealistic cycles | Overly synchronous updating scheme. | Switch from synchronous to an asynchronous updating scheme (e.g., ARBNs or DARBNs [76]), which more realistically captures biological timing. |
| PDE solver is too slow | Fine spatial grid; High-dimensional domain; Complex geometry. | Use coarser grid for initial exploration; employ operator learning surrogates [74]; leverage high-performance computing (HPC). |
| Model predictions lack biological insight | Model is a "black box"; Over-reliance on data-fitting without mechanistic understanding. | Adopt Scientific Machine Learning (SciML) [73]: integrate the mechanistic model with ML to open the "black box" and ensure predictions are physiologically interpretable. |
Section 3: Experimental Protocols and Workflows
Protocol: Converting an ODE Model to a Boolean Network
This protocol is useful for gaining topological insights when ODE parameters are unknown or to analyze large-scale dynamics [72].
- Step 0: Preliminaries. Start with a normalized ODE system ( y′(t)=f(y) ), where the right-hand side consists of sums and products of monotone functions (e.g., Hill functions, mass action). Ensure variables are normalized to [0,1] [72].
- Step 1: Discretization. Perform an Euler-like discretization. For a variable ( yi ), the update rule is derived from the sign of the right-hand side of its ODE: ( yi(t+1) = 1 ) if ( fi(y(t)) > 0 ), and ( yi(t+1) = 0 ) if ( f_i(y(t)) < 0 ). This translates the continuous rate of change into a discrete ON/OFF decision.
- Step 2: Define Logic Gates. Convert the mathematical relationships in ( f_i(y) ) into logical Boolean operators (AND, OR, NOT). For example, a synthesis term that requires two activators becomes an AND gate; a term with multiple independent synthesis paths becomes an OR gate.
- Step 3: Validation. Compare the dynamics of the resulting Boolean network with the original ODE model or qualitative biological data. Check if steady states (attractors) and key behaviors (e.g., oscillations) are conserved [72].
- Step 4: Analysis. Use the Boolean model for tasks that are difficult with ODEs, such as a global attractor analysis or exhaustive in-silico knock-out studies to identify critical network components.
Diagram: Workflow for converting an ODE model to a Boolean network.
Protocol: Implementing a Physics-Informed DeepONet for PDEs
This protocol outlines the use of a Physics-Informed DeepONet to create a fast surrogate for a costly PDE solver, ideal for parameter estimation and uncertainty quantification in spatial biological models [77].
- Problem Formulation: Define your PDE with initial and boundary conditions, identifying the input function ( v ) (e.g., a spatially varying parameter) and the output solution ( u ) you wish to learn.
- Data Generation:
- Generate a set of ( N ) input functions ( {vi(x)} ).
- Use a high-fidelity numerical solver (e.g., Finite Element Method) to compute the solutions ( {ui(t, x)} ) for each input. Alternatively, for a fully physics-informed approach, this step can be skipped, and the PDE itself will act as the supervisor.
- Network Architecture:
- Branch Net: Takes the discretized values of the input function ( v ) as input.
- Trunk Net: Takes the spatial (( x )) and temporal (( t )) coordinates as input.
- The outputs of the branch and trunk nets are combined via a dot product to generate the predicted solution ( u(t, x) ) [77].
- Training:
- Data Loss: Minimize the mean-squared error between predictions and high-fidelity solution data (if used).
- Physics Loss: Calculate the PDE residual using automatic differentiation on the network's output and minimize it. This ensures the network's predictions obey the physical laws encoded in the PDE, even without data.
- Inference: Use the trained model to instantly predict the full solution ( u(t, x) ) for any new input function ( v_{new} ) without solving the PDE numerically.
Diagram: Physics-Informed DeepONet architecture for solving PDEs.
Section 4: The Scientist's Toolkit
Research Reagent Solutions
Table 3: Essential computational tools and resources for modeling.
| Item | Function | Example Uses |
|---|---|---|
| DifferentialEquations.jl (Julia) | A unified suite for performing ODE/PDE solving with a wide range of high-performance solvers [78]. | Solving stiff/non-stiff ODEs of immune dynamics; parameter estimation. |
| Logic Modeling Software (e.g., BooINet, GINsim) | Software specifically designed for building, simulating, and analyzing Boolean networks. | Identifying attractors in cell signaling networks; simulating knock-out experiments [75]. |
| Physics-Informed DeepONet Framework | A neural network framework for learning solution operators of differential equations [77]. | Building fast surrogates for expensive spatial biological models (e.g., granuloma formation). |
| NODE-ONet Framework | An encoder-neural ODE-decoder framework for learning dynamics of PDEs with good generalization [74]. | Predicting long-term behavior of reaction-diffusion systems in tissue beyond the trained time frame. |
| Sensitivity & Identifiability Analysis Tools | Software to quantify how model outputs depend on parameters. | Pinpointing unidentifiable parameters in a complex ODE model of cytokine crosstalk. |
ODE Solver Selection Guide
Table 4: Recommended ODE solvers for different problem types, based on DifferentialEquations.jl [78].
| Problem Type | Recommended Solver(s) | Key Characteristics |
|---|---|---|
| Non-Stiff Problems (Default) | Tsit5(), BS5() |
Efficient and accurate for most problems; good general-purpose choice. |
| Stiff Problems (Low Accuracy) | Rosenbrock23(), TRBDF2() |
Robust to stiffness and oscillations; suitable for tolerances >1e-2. |
| Stiff Problems (Medium/High Accuracy) | Rodas5P(), KenCarp4() |
Efficient and reliable for tolerances from ~1e-8 to 1e-2; handles nonlinear parabolic PDE discretizations well. |
| Very Large Systems (>1000 ODEs) | QNDF(), FBDF() |
Efficient for large systems where Jacobian factorization is costly; minimal oscillations. |
| Unknown Stiffness | AutoTsit5(Rosenbrock23()) |
Automatically detects stiffness and switches between non-stiff and stiff solvers. |
Validating Predictions Against Experimental Data from Mouse Peritonitis Models
Frequently Asked Questions (FAQs)
Q1: What are the most common sources of uncertainty when fitting a mathematical model to data from mouse peritonitis experiments? Uncertainty in parameter estimates primarily stems from two key areas: the model's structure and the available data. Structural identifiability is a fundamental property that determines if a model's unknown parameters can be uniquely determined from perfect, noise-free data. If parameters are correlated, the model may be structurally unidentifiable. Practical identifiability concerns whether parameters can be accurately estimated given the constraints of real-world data, which is often limited in frequency, noisy, and may not cover all model variables [14].
Q2: Our model simulations do not match the biphasic decay of inflammatory markers seen in our experimental data. What model features might be missing? A biphasic decay often indicates the involvement of adaptive immune responses not captured in simpler models. A basic model might only include target cells, infected cells, and virus (or pathogens). To capture biphasic dynamics, you may need to incorporate:
- Immune Effector Cells: Explicitly include a variable for immune cells like CD8+ T cells. Their recruitment and activity, often following a density-dependent function (e.g., ( \frac{\deltaE}{K\delta + I} )), can drive the second phase of decay [14].
- Time Delays: Account for the delay between infection and the expansion of the adaptive immune response using a delay differential term, such as ( \eta E I2(t-\tauI) ) [14].
- Non-linear Clearing: Instead of a constant infected cell death rate, use a term like ( \frac{\delta}{K_\delta + I}I ) that increases as the population of infected cells decreases [14].
Q3: How can we determine if our model is too complex for the experimental data we have collected? Perform a practical identifiability analysis. After ensuring your model is structurally identifiable, estimate parameters from your noisy, limited data (e.g., daily measurements). Then, analyze the confidence intervals of the parameter estimates. Parameters with very large confidence intervals are practically unidentifiable with your current data. This indicates a need for more frequent data points, measurements of additional model variables (e.g., immune cell counts alongside pathogen titers), or a model simplification [14].
Q4: What experimental readouts are most critical for validating a comprehensive mathematical model of LPS-induced peritonitis? To constrain a complex model, multiple types of data are essential. Key quantitative readouts include:
- Cytokine Concentrations: Measure levels of key pro-inflammatory (TNF-α, IL-6, IL-1β) and anti-inflammatory (IL-10) cytokines in peritoneal lavage fluid or plasma over time [8] [79].
- Pathogen Load: For infectious models, viral or bacterial titers are fundamental [14].
- Immune Cell Populations: Flow cytometry data for immune cells like neutrophils, macrophages, and CD8+ T cells in the peritoneum or blood [14] [80].
- Clinical Signs: Body temperature, heart rate, and blood pressure provide systemic validation of the model's physiological impact [8].
- Transcriptomic Data: RNA sequencing of peritoneal tissue or blood can identify hub genes and pathways, providing a molecular-level validation target [80] [79].
Q5: How can molecular dynamics simulations be relevant to my mathematical model of inflammation? Molecular dynamics (MD) simulations bridge the gap between transcriptomic data and protein function. While your mathematical model operates at a cellular/organism level, MD simulations can:
- Validate Hub Genes: Provide optimized 3D structures for proteins encoded by hub genes identified in transcriptomic studies (e.g., from RNA-seq of peritoneal tissue) [80] [79].
- Inform Model Mechanisms: Simulate how specific inhibitors (e.g., TAK-242, a TLR4 inhibitor) bind to and alter the conformation of their target proteins, providing a mechanistic basis for including certain interactions or drug effects in your larger-scale model [79].
Troubleshooting Guides
Issue 1: Poor Model Fit to Time-Course Data
Problem: Your model simulations consistently deviate from the experimental time-course data for key variables like cytokine concentration.
Solution Steps:
- Verify Structural Identifiability: Before fitting, perform a structural identifiability analysis on your model. This confirms that, in theory, all parameters can be uniquely identified from perfect data [14].
- Check Practical Identifiability: If the model is structurally identifiable but fits poorly, perform a practical identifiability analysis using your specific dataset. This will reveal if the issue is insufficient or noisy data [14].
- Review Model Mechanisms:
- Acute vs. Prolonged Stimulus: Ensure your model is suited for your experiment. A model designed for a short LPS bolus may not perform well under continuous LPS infusion [8].
- Include Critical Feedback: Models lacking key anti-inflammatory feedback loops (e.g., via IL-10) may become unstable or fail to capture resolution dynamics. Ensure your model includes these essential regulatory interactions [8].
Issue 2: Experimentally Observed Pathway Inhibition Does Not Match Model Predictions
Problem: You administer an inhibitor (e.g., TAK-242 for TLR4) and the experimentally measured cytokine response is significantly different from your model's prediction.
Solution Steps:
- Re-calibrate with Inhibition Data: Incorporate the experimental data from the inhibition experiment into your model calibration process. This helps re-estimate parameters related to the inhibited pathway.
- Refine the Inhibition Mechanism: The initial model might oversimplify the inhibition. Use molecular dynamics and docking studies to understand the precise binding mode and efficacy of the inhibitor, which can inform a more accurate mathematical representation of the inhibition in your model [79].
- Check for Compensatory Pathways: The model may lack alternative (compensatory) signaling pathways that become active when the primary pathway is blocked. Transcriptomic data from inhibited versus non-inhibited conditions can help identify these alternative routes for inclusion in the model [79].
Issue 3: Inability to Reproduce a Biphasic Immune Response
Problem: Your model shows a simple, monophasic response, but your in vivo data clearly shows two distinct phases of immune activation and resolution.
Solution Steps:
- Model Selection: Start with a basic model and progressively add complexity. The table below outlines the evolution of model features that can help capture biphasic dynamics.
Table: Evolution of Within-Host Models to Capture Complex Dynamics
| Model Name | Key Features | Typical Data Used | Ability to Capture Biphasic Decay |
|---|---|---|---|
| Basic Target Cell | Target cells (T), infected cells (I), virus/pathogen (V) [14] | Virus titer data [14] | Limited |
| With Eclipse Phase | Adds eclipse phase (Iâ‚, Iâ‚‚); delay in viral production [14] | Virus titer data [14] | Improved with non-linear clearing [14] |
| With Adaptive Immunity | Explicitly includes immune effector cells (E); time-delayed expansion; cell killing [14] | Virus titer + immune cell data [14] | Yes, can explicitly model the second phase driven by adaptive immunity [14] |
- Incorporate Innate-to-Adaptive Switch: Ensure your model includes a mechanism for the innate immune response to activate the adaptive immune system. This often requires a DDE to account for the time needed for T-cell priming and expansion [14].
- Validate with Additional Data: Fit the extended model not just to pathogen load, but also to time-course data for immune cell populations (e.g., CD8+ T cells). This provides critical constraints for the new parameters [14].
Experimental Protocols & Data
Key Experimental Model: LPS-Induced Murine Peritonitis
This well-established model is used to study the acute inflammatory response and is a key source of data for model validation.
Detailed Methodology:
- Animals: Use male C57BL/6 mice, 6–8 weeks old [80].
- Induction of Peritonitis: Administer a single intraperitoneal (i.p.) injection of LPS (e.g., 10 mg/kg) dissolved in saline [79].
- Intervention Studies: To test specific mechanisms, pre-treat or co-treat with compounds like the TLR4 inhibitor TAK-242 (e.g., 3 mg/kg, administered intravenously) [79].
- Sample Collection:
- Peritoneal Lavage Fluid: At designated time points, lavage the peritoneal cavity with saline. Collect the fluid and centrifuge to separate cells from supernatant [79].
- Blood: Collect via cardiac puncture or other methods for plasma and transcriptomic analysis [80].
- Tissue: Isolve peritoneal tissue for transcriptomic sequencing [79].
- Data Generation:
- Cytokine Measurement: Use ELISA kits to quantify cytokines (TNF-α, IL-6, IL-1β, IFN-γ) and other mediators like nitric oxide in the lavage supernatant [79].
- Transcriptomics: Isolate RNA from blood or tissue and perform bulk RNA-sequencing. Subsequent bioinformatics analysis includes identifying Differentially Expressed Genes (DEGs), pathway enrichment, and hub gene analysis [80] [79].
Table: Example Quantitative Data from LPS-Induced Murine Peritonitis Model (Cytokines in Peritoneal Lavage Fluid)
| Cytokine / Mediator | Control Group | LPS Group | LPS + TAK-242 Group | Measurement Method |
|---|---|---|---|---|
| TNF-α (pg/mL) | 47.09 ± 13.01 | 546.7 ± 156.0 | 312.6 ± 73.53 | ELISA [79] |
| IL-6 (pg/mL) | 23.89 ± 6.485 | 403.4 ± 42.08 | 180.2 ± 30.09 | ELISA [79] |
| IL-1β (pg/mL) | 8.345 ± 2.746 | 921.7 ± 114.9 | 400.2 ± 59.19 | ELISA [79] |
| IFN-γ (pg/mL) | 11.61 ± 4.252 | 570.6 ± 65.84 | 303.4 ± 30.20 | ELISA [79] |
| Nitric Oxide (μg/mL) | 3.942 ± 0.242 | 14.47 ± 0.248 | 10.81 ± 0.722 | Assay Kit [79] |
The Scientist's Toolkit: Research Reagent Solutions
Table: Essential Materials for Mouse Peritonitis and Validation Studies
| Item | Function/Application | Example |
|---|---|---|
| TLR4 Agonist | Induces sterile inflammation via the TLR4 pathway; core of the peritonitis model. | Lipopolysaccharide (LPS) [79] |
| TLR4 Inhibitor | Tool for mechanistic validation; blocks the TLR4 pathway to test model predictions. | TAK-242 (Resatorvid) [79] |
| ELISA Kits | Quantify protein levels of cytokines and chemokines in biological fluids. | TNF-α, IL-6, IL-1β, IL-10 ELISA kits [8] [79] |
| RNA-seq Kit | Profile genome-wide gene expression changes in tissue or blood samples. | Bulk RNA-sequencing services/reagents [80] [79] |
| Molecular Dynamics Software | Simulate protein structure and dynamics; validate hub gene function. | GROMACS, AMBER, NAMD [80] [79] |
Workflow and Pathway Visualizations
Assessing Predictive Power for Therapeutic Interventions and Patient Stratification
Troubleshooting Guides and FAQs
Section 1: Mathematical Model Development and Identifiability
FAQ: What are the most common causes of identifiability issues in mathematical models of inflammation? Identifiability issues primarily arise when multiple parameter combinations produce identical model outputs, making unique parameter estimation impossible. Common causes include: (1) Over-parameterization - too many parameters for the available data; (2) Insufficient data - lack of temporal or component-specific measurements; (3) Correlated parameters - parameters that have similar effects on model outputs; (4) Poor experimental design - data that doesn't sufficiently excite system dynamics.
Troubleshooting Guide: Resolving Structural Non-Identifiability
- Problem: Model outputs are insensitive to certain parameters regardless of data quality.
- Diagnosis: Perform structural identifiability analysis using symbolic computation before parameter estimation.
- Solution:
- Simplify model structure by reducing biologically redundant parameters.
- Reparameterize groups of non-identifiable parameters into identifiable combinations.
- Incorporate prior knowledge from literature to fix certain parameter values.
- Reference: Profile likelihood analysis (PLA) has been successfully applied to inflammation models to confirm unique parameter estimation is possible with available data [8].
Troubleshooting Guide: Addressing Practical Non-Identifiability
- Problem: Parameters cannot be uniquely identified due to limited or noisy data.
- Diagnosis: Local sensitivity analysis reveals low sensitivity indices for certain parameters.
- Solution:
- Design new experiments targeting specific timepoints or components where sensitivity is highest.
- Increase data frequency during critical transition phases (e.g., pro-inflammatory to anti-inflammatory shift).
- Utilize multi-scale data (molecular, cellular, organ-level) to constrain parameters.
- Reference: In a sepsis inflammation model, sensitivity analysis identified six key parameters (including cytokine scaling factors and mRNA half-lives) that most significantly influenced model outputs and were suitable for estimation [8].
Section 2: Model Validation and Predictive Power Assessment
FAQ: How do I validate that my model has genuine predictive power for therapeutic interventions? True predictive validation requires: (1) External validation - testing model predictions on completely independent datasets not used for model training/calibration; (2) Prospective validation - making predictions before experimental results are known; (3) Interventional validation - accurately predicting outcomes of therapeutic perturbations not present in training data.
Troubleshooting Guide: Improving Model Predictive Performance
- Problem: Model fits calibration data well but fails to predict new experimental outcomes.
- Diagnosis: Model may be overfitted to specific conditions or lack key biological mechanisms.
- Solution:
- Implement cross-validation techniques to detect overfitting.
- Incorporate additional biological constraints based on known physiology.
- Use ensemble modeling approaches to capture system variability.
- Validate against multiple endpoints (e.g., cytokine dynamics, cell counts, clinical outcomes).
- Reference: A modular immune response model for COVID-19 was validated by testing its ability to reproduce biologically relevant behaviors under various conditions, including immunity hyperactivation and co-infection scenarios [30].
Experimental Protocol: Prospective Validation for Patient Stratification
- Pre-specification: Before analysis, define stratification criteria, primary endpoints, and statistical methods.
- Blinding: Keep treatment assignments blinded during model application to prevent bias.
- External Cohort: Apply trained model to a completely independent patient cohort.
- Endpoint Assessment: Compare predicted vs. actual clinical outcomes across stratified groups.
- Efficiency Metrics: Calculate sample size reductions achievable through stratification.
Table 1: Performance Metrics from AI-Guided Stratification in Alzheimer's Trial [81]
| Metric | Standard Approach | AI-Guided PPM Stratification | Improvement |
|---|---|---|---|
| Classification Accuracy | Not Applicable | 91.1% | Baseline |
| Sensitivity | Not Applicable | 87.5% | Baseline |
| Specificity | Not Applicable | 94.2% | Baseline |
| Treatment Effect (CDR-SOB) | Non-significant | 46% slowing of decline | Clinically significant effect demonstrated |
| Sample Size Requirements | Larger reference group | Substantially decreased | Enhanced trial efficiency |
Section 3: AI Integration and Computational Methods
FAQ: What are the key considerations when integrating AI with mechanistic models for patient stratification? Successful integration requires: (1) Interpretability - AI components should provide insight into biological mechanisms; (2) Validation - rigorous testing on independent clinical datasets; (3) Clinical relevance - stratification should align with biologically meaningful subgroups; (4) Regulatory compliance - documentation for clinical trial applications.
Troubleshooting Guide: Addressing "Black Box" Limitations in AI Models
- Problem: AI model provides accurate predictions but no insight into biological mechanisms.
- Diagnosis: Model uses complex architectures without interpretability features.
- Solution:
- Implement interpretability methods like SHAP (Shapley Additive Explanations) to quantify feature importance.
- Use inherently interpretable models like Generalized Metric Learning Vector Quantization (GMLVQ).
- Interrogate model components (e.g., prototype analysis in GMLVQ) to understand class representations.
- Reference: The PPM model for Alzheimer's stratification used GMLVQ's interpretable architecture to identify β-amyloid burden as the most discriminative feature, consistent with known biology [81].
Experimental Protocol: Developing an Interpretable Predictive Model
- Feature Selection: Incorporate multimodal data (clinical, imaging, molecular biomarkers).
- Model Training: Use interpretable architectures (e.g., GMLVQ) that provide prototype analysis.
- Validation: Test on independent datasets against longitudinal clinical outcomes.
- Mechanistic Interpretation: Interrogate metric tensors to understand feature contributions and interactions.
- Clinical Application: Generate prognostic indices for stratification and outcome prediction.
Table 2: Comparison of Computational Approaches for Inflammation Research [82] [83] [84]
| Method | Primary Application | Key Features | Validation Approach |
|---|---|---|---|
| PreAIP Predictor [84] | Anti-inflammatory peptide prediction | Integrates multiple complementary features (sequence, evolutionary, structural) | 10-fold cross-validation (AUC: 0.833) |
| VC-SEPS Algorithm [83] | Early sepsis prediction | Deep learning on EMR data; provides risk scores | Prospective validation on 6,455 patients (AUROC: 0.880) |
| Inflammation Indices Model [82] | Identify therapeutic targets | Sensitivity and correlation analysis of timing/amount indices | Simulation of thousands of inflammatory scenarios |
| Modular Immune Model [30] | SARS-CoV-2 infection dynamics | Multi-scale, multi-compartment; integrates innate/adaptive immunity | Parameter optimization against experimental data |
Section 4: Data Integration and Multi-Scale Modeling
FAQ: How can I effectively integrate data across multiple biological scales in inflammation models? Effective multi-scale integration requires: (1) Modular design - creating interchangeable model components for different biological scales; (2) Data standardization - establishing consistent formats and units across experimental sources; (3) Scale-specific validation - verifying model performance at each biological scale independently; (4) Efficient parameter estimation - using hierarchical methods that leverage information across scales.
Troubleshooting Guide: Managing Computational Complexity in Multi-Scale Models
- Problem: Model becomes computationally intractable with too many scales and components.
- Diagnosis: Simulation times are excessive, parameter estimation is unstable.
- Solution:
- Implement modular framework allowing components to run at different temporal/spatial resolutions.
- Use model reduction techniques for less critical processes.
- Employ parallel computing for parameter estimation and sensitivity analysis.
- Establish clear communication protocols between model modules.
- Reference: A modular mathematical model of immune response to SARS-CoV-2 was successfully developed by integrating manually reviewed and validated models of different infections, creating a multi-compartmental framework [30].
Model Development Workflow
Inflammation Resolution Pathway
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Computational Tools and Resources for Inflammation Modeling
| Tool/Resource | Type | Primary Function | Application Example |
|---|---|---|---|
| Profile Likelihood Analysis | Statistical Method | Assess parameter identifiability | Determining which parameters can be uniquely estimated from data [8] |
| GMLVQ Algorithm | Machine Learning | Interpretable classification and stratification | Patient stratification in Alzheimer's trials [81] |
| SHAP Analysis | Model Interpretation | Explain AI model predictions | Feature importance analysis in sepsis prediction models [83] |
| BioUML Platform | Modeling Environment | Multi-scale model development and simulation | Modular immune response model for COVID-19 [30] |
| Inflammation Indices | Quantitative Metrics | Characterize timing and intensity of response | Ψmax, Tact, Ri, Rp for neutrophil/macrophage trajectories [82] |
| Digital Twin Framework | Personalized Modeling | Patient-specific simulation platform | Immune Digital Twin paradigm for personalized therapy [30] |
Conclusion
Resolving identifiability issues is not merely a technical exercise but a fundamental prerequisite for developing mathematically rigorous and biologically meaningful models of inflammation. A systematic approach—combining foundational understanding, robust methodological toolkits, strategic troubleshooting, and rigorous validation—is essential to transform non-identifiable models into reliable tools for discovery. Future progress hinges on the adoption of standardized identifiability analysis pipelines, the development of more accessible software, and closer integration of modeling with targeted experimental design. For biomedical and clinical research, overcoming these challenges is the key to unlocking the full potential of mathematical models in predicting patient-specific outcomes, optimizing therapeutic interventions, and ultimately guiding the development of novel treatments for complex inflammatory diseases.
References
- 1. On structural and practical identifiability [https://www.sciencedirect.com/science/article/pii/S245231002100007X]
- 2. Structural and practical identifiability analysis in bioengineering [https://jbioleng.biomedcentral.com/articles/10.1186/s13036-024-00410-x]
- 3. Structural identifiability [https://en.wikipedia.org/wiki/Structural_identifiability]
- 4. Navigating the landscape of parameter identifiability methods [https://pmc.ncbi.nlm.nih.gov/articles/PMC11247124/]
- 5. Practical Identifiability in Modeling [https://www.emergentmind.com/topics/practical-identifiability]
- 6. How do we Deal with Identifiability Problems in Statistics? [https://stats.stackexchange.com/questions/620217/how-do-we-deal-with-identifiability-problems-in-statistics]
- 7. Structural and Practical Identifiability of Phenomenological ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12031297/]
- 8. Mathematical model of the inflammatory response to acute ... [https://www.nature.com/articles/s41540-024-00473-y]
- 9. Mathematical modelling of inflammatory process and obesity ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0323258]
- 10. Identifiability of basic models and parameters in ... [https://www.degruyterbrill.com/document/doi/10.1515/rnam-2025-0015/html?srsltid=AfmBOoqg3JC1QvGnJvYm8aFYwAaR81K0YJe2AaRu1f2_JYk9tcr0I2qX]
- 11. Identifiability in epidemic models with prior immunity and ... [https://arxiv.org/abs/2506.07825]
- 12. Identifiability Challenges in Mathematical Models of Viral ... [https://www.sciencedirect.com/science/article/pii/S2405896315027597]
- 13. A network approach to define the predictive role of immune ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2023.1199089/full]
- 14. Identifiability investigation of within-host models of acute ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11100786/]
- 15. Predictive power of non-identifiable models [https://www.nature.com/articles/s41598-023-37939-8]
- 16. Nonidentifiability in model calibration and implications for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6156799/]
- 17. Identifiability methods for biological systems: Determining ... [https://www.sciencedirect.com/science/article/pii/S0098135424001017]
- 18. A Tutorial on Structural Identifiability of Epidemic Models ... [https://arxiv.org/html/2505.10517v1]
- 19. Systemic Inflammation: Methodological Approaches to ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0155138]
- 20. Identifiability of parameters in mathematical models ... [https://www.nature.com/articles/s41598-022-18683-x]
- 21. A Systematic Computational Framework for Practical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12463036/]
- 22. Identifiability of basic models and parameters in ... [https://www.degruyterbrill.com/document/doi/10.1515/rnam-2025-0015/html?srsltid=AfmBOorXd1tMvzFW-h5WNdY-IQDZnKQmZvTr_w5yEc1aiATSxRXzdPMH]
- 23. Chronic LCMV infection regulates the effector T cell ... [https://www.nature.com/articles/s12276-023-00991-5]
- 24. Structural analysis of differential-algebraic equation ... [https://www.sciencedirect.com/science/article/pii/0098135494000945]
- 25. Laplace transform [https://en.wikipedia.org/wiki/Laplace_transform]
- 26. Applications of Laplace Transform in Engineering [https://www.linkedin.com/pulse/applications-laplace-transform-engineering-comprehensive-sathiyaraj-g]
- 27. SciML/StructuralIdentifiability.jl: Fast and automatic ... [https://github.com/SciML/StructuralIdentifiability.jl]
- 28. Structural Identifiability Analysis · Catalyst.jl [https://docs.sciml.ai/Catalyst/v15.0/inverse_problems/structural_identifiability/]
- 29. Reduced order modeling and analysis of the human ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0187373]
- 30. A Modular Mathematical Model of the Immune Response for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12115727/]
- 31. A Whole-Body Mathematical Model of Sepsis Progression ... [https://www.frontiersin.org/journals/physiology/articles/10.3389/fphys.2019.01321/full]
- 32. A Tutorial on Structural Identifiability of Epidemic Models ... [https://arxiv.org/abs/2505.10517]
- 33. of a Global of Acute... Sensitivity Analysis Mathematical Model [https://pmc.ncbi.nlm.nih.gov/articles/PMC4125477/]
- 34. Optimizing Genetic Circuits by Global Sensitivity Analysis [https://pmc.ncbi.nlm.nih.gov/articles/PMC1304645/]
- 35. GUI–HDMR – A software tool for global sensitivity analysis ... [https://www.sciencedirect.com/science/article/abs/pii/S1364815208002168]
- 36. for fiber reinforced composite fiber path... Global sensitivity analysis [https://link.springer.com/article/10.1007/s00158-017-1681-9]
- 37. Mathematical modeling of inflammatory response in ... [https://hammer.purdue.edu/articles/thesis/_b_Mathematical_modeling_of_inflammatory_response_in_mammalian_macrophages_using_cybernetic_framework_and_novel_information-theoretic_approaches_b_/26278450]
- 38. Troubleshooting [https://turinglang.org/docs/usage/troubleshooting/]
- 39. Hamiltonian Monte Carlo gets stuck at tiny local maxima [https://discourse.julialang.org/t/hamiltonian-monte-carlo-gets-stuck-at-tiny-local-maxima/108389]
- 40. Symplectic integration and dynamic Hamiltonian Monte Carlo [https://discourse.julialang.org/t/symplectic-integration-and-dynamic-hamiltonian-monte-carlo/14602]
- 41. Frequently Asked Questions [https://turinglang.org/docs/faq/]
- 42. Introduction · DynamicHMC.jl [https://www.tamaspapp.eu/DynamicHMC.jl/stable/]
- 43. tpapp/DynamicHMC.jl: Implementation of robust dynamic ... [https://github.com/tpapp/DynamicHMC.jl]
- 44. Robust parameter estimation and identifiability analysis ... [https://www.nature.com/articles/s41540-024-00460-3]
- 45. Jaxkineticmodel: Neural ordinary differential equations ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12258584/]
- 46. Improving generalization ability of deep-learning-based ... [https://www.nature.com/articles/s44387-025-00026-6]
- 47. Low-dimensional neural ODEs and their application in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10982100/]
- 48. Identifiability, indistinguishability, and other problems in ... [https://www.imsi.institute/videos/identifiability-indistinguishability-and-other-problems-in-biological-modeling/]
- 49. Reparameterization trick [https://en.wikipedia.org/wiki/Reparameterization_trick]
- 50. Understanding the Reparameterization Trick [https://medium.com/@ml_dl_explained/understanding-the-reparameterization-trick-be349756b91b]
- 51. FAQ [https://graphviz.org/faq/]
- 52. Change font attributes within a (node) label's text? - Help [https://forum.graphviz.org/t/change-font-attributes-within-a-node-labels-text/779]
- 53. graphviz nodes of different colors [https://stackoverflow.com/questions/13106328/graphviz-nodes-of-different-colors]
- 54. colorscheme [https://graphviz.org/docs/attrs/colorscheme/]
- 55. Lipid droplet-targeting carbonized polymer dots for ... [https://www.sciencedirect.com/science/article/abs/pii/S0003267025001849]
- 56. Introductory overview of identifiability analysis [https://www.sciencedirect.com/science/article/abs/pii/S1364815218307278]
- 57. Mathematical Modeling of Neuroinflammation in ... [https://pubmed.ncbi.nlm.nih.gov/40801430/]
- 58. Identifiability of basic models and parameters in ... [https://www.degruyterbrill.com/document/doi/10.1515/rnam-2025-0015/html?srsltid=AfmBOooV_NgSASMCDlZIHF945SRoWor5d6HfmKEll9ZxBPYMYbwcfnxV]
- 59. Parameter identifiability and model selection for partial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10914513/]
- 60. A simple in-host model for COVID-19 with treatments [https://link.springer.com/article/10.1007/s00285-022-01849-6]
- 61. Review of Mathematical Modeling of the Inflammatory ... [https://www.frontiersin.org/journals/applied-mathematics-and-statistics/articles/10.3389/fams.2020.00036/full]
- 62. How to be a Better Troubleshooter in Your Laboratory [https://www.goldbio.com/blogs/articles/how-to-be-a-better-troubleshooter?srsltid=AfmBOoq1U92lblOF9CIrVaTqHN0ujGPMzufBD8OmpeupyDEJqrawixtn]
- 63. 5 Steps to Follow for Troubleshooting Experiments [https://blog.edvotek.com/2022/06/21/5-steps-to-follow-for-troubleshooting-experiments/]
- 64. Local and systemic factors in the resolution of inflammation [https://www.sciencedirect.com/science/article/pii/S0022519325002541]
- 65. Development of a mathematical model of the innate ... [https://www.sciencedirect.com/science/article/abs/pii/S0022519325001250]
- 66. On-chip 3D potency assay for prediction of clinical ... [https://www.nature.com/articles/s41467-025-60158-w]
- 67. Mendelian randomization integrated with multi-omics ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1678444/full]
- 68. Single-cell multi-omics-based immune temporal network ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12375439/]
- 69. Increased levels of ficolin-1 and of C3dg are independently ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12521454/]
- 70. Age-related inflammatory changes and perineuronal net ... [https://jneuroinflammation.biomedcentral.com/articles/10.1186/s12974-025-03568-3]
- 71. A Multi-Platform Approach to Small Molecule Anti-IBD Drug ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12566827/]
- 72. Complementing ODE-Based System Analysis Using Boolean ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4619740/]
- 73. The rise of scientific machine learning: a perspective on ... [https://www.frontiersin.org/journals/systems-biology/articles/10.3389/fsysb.2024.1407994/full]
- 74. Deep Neural ODE Operator Networks for PDEs [https://arxiv.org/html/2510.15651v1]
- 75. Logic-Based Models for the Analysis of Cell Signaling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2853906/]
- 76. Modelling brain dynamics by Boolean networks [https://www.nature.com/articles/s41598-022-20979-x]
- 77. Operator Learning via Physics-Informed DeepONet [https://towardsdatascience.com/operator-learning-via-physics-informed-deeponet-lets-implement-it-from-scratch-6659f3179887/]
- 78. ODE Solvers · DifferentialEquations.jl [https://docs.sciml.ai/DiffEqDocs/stable/solvers/ode_solve/]
- 79. Transcriptomics Changes in the Peritoneum of Mice with ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8657704/]
- 80. Insights into transcriptomic changes in blood of a mouse ... [https://www.sciencedirect.com/science/article/abs/pii/S0041008X25003990]
- 81. AI-guided patient stratification improves outcomes and ... [https://www.nature.com/articles/s41467-025-61355-3]
- 82. Computational Identification of Mechanistic Factors That ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4669096/]
- 83. Validation of an artificial intelligence-based algorithm for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12182025/]
- 84. PreAIP: Computational Prediction of Anti-inflammatory ... [https://www.frontiersin.org/journals/genetics/articles/10.3389/fgene.2019.00129/full]